DeepMIB - Directories and Preprocessing tab
This tab allows choosing directories with images for training and prediction as well as various parameters used during image loading and preprocessing
Back to Index --> User Guide --> Menu --> Tools Menu --> Deep learning segmentation
Contents
-
Widgets and settings -
Preprocessing of files -
Organization of directories without preprocessing for semantic segmentation -
Organization of directories with preprocessing for semantic segmentation -
Organization of directories for 2D patch-wise workflow
Widgets and settings
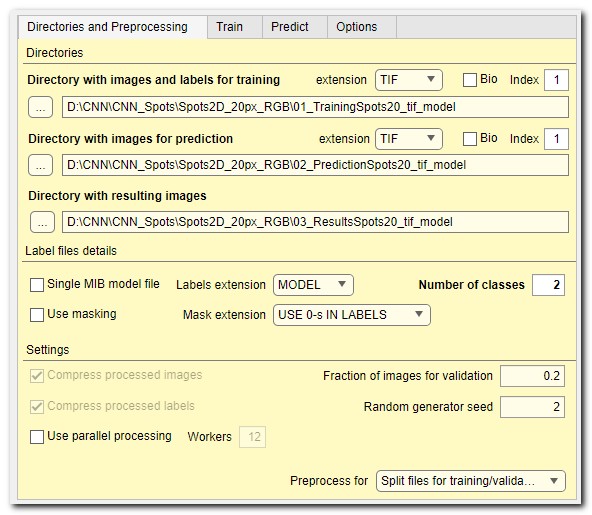
Directory with images and labels for training
[ used only for training ]
use these widgets to select directory that contain images and model to be
used for training. For the organization of directories see the
organization schemes below.
For 2D networks the files should contain individual 2D images, while for 3D networks
individual 3D datasets.
The extension ▼ dropdown menu on the right-hand side can be used to specify extension
of the image files.
The [✓] Bio checkbox toggles standard or Bio-format readers for loading images.
If the Bio-Format file is a collection of image, the Index... edit box can be used
to specify an index of the file within the container.
For better performance, it is recommended to convert Bio-Formats compatible images to standard formats or to use the Preprocessing option (see below).
Important notes considering training files
- Number of model or mask files should match the number of image files
(one exception is 2D networks, where it is allowed to have a single
model file in MIB *.model format, when Single MIB model file:
ticked). This option requires
data preprocessing - For labels in standard image formats it is important to specify number of classes including the Exterior into the Number of classes edit box
- Important! It is not possible to use numbers as names of materials, please name materials in a sensible way when using the *.model format!
Directory with images for prediction
[ used only for prediction ]
use these widgets to specify directory with images for prediction (named 2_Prediction in
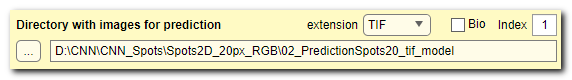
The image files should be placed under Images subfolder
(it is also possible to place images directly into a folder specified in this panel).
Optionally, when the ground truth labels for prediction images are available, they can be placed under Labels subfolder.
When the preprocessing mode is used the images from this folder are
converted and saved to 3_Results\Prediction images directory.
When the ground truth labels are present, they are also processed and copied to
3_Results\PredictionImages\GroundTruthLabels. These labels can
be used for evaluation of results (see
For 2D networks the files should contain individual 2D images or 3D stacks, while for 3D networks
individual 3D datasets.
The extension ▼ dropdown menu on the right-hand side can be used to specify extension
of the image files. The [✓] Bio checkbox toggles standard or Bio-format readers for loading the images.
If the Bio-Format file is a collection of image, the Index edit box can be used
to specify an index of the file within the container.
Directory with resulting images
use these widgets to specify the main output directory; results and all preprocessed images are stored there.
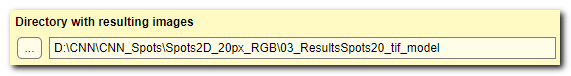
All subfolders inside this directory are automatically created by Deep MIB:
Description of directories created by DeepMIB
- PredictionImages, place for the prepocessed images for prediction
- PredictionImages\GroundTruthLabels, place for ground truth labels for prediction images, when available
- PredictionImages\ResultsModels, the main outout directory with generated labels after prediction. The 2D models can be combined in MIB by selecting the files using the ⇧ Shift+left mouse click during loading
- PredictionImages\ResultsScores, folder for generated prediction scores (probability) for each material. The score values are scaled between 0 and 255
- ScoreNetwork, for accuracy and loss score plots, when the Export training plots option of the Train tab is ticked and for storing checkpoints of the network after each epoch (or specified frequency), when the [✓] Save progress after each epoch checkbox is ticked. The score files are started with a date-time tag and overwritten when a new training is started
- TrainImages, images to be used for training (only for preprocessing mode)
- TrainLabels, labels accompanying images to be used for training (only for preprocessing mode)
- ValidationImages, images to be used for validation during training (only for preprocessing mode)
- ValidationLabels, labels accompanying images for validation (only for preprocessing mode)
Label file details
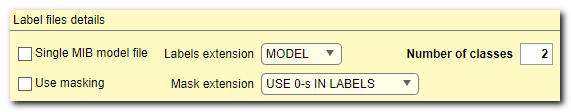
- The [✓] Single MIB model file checkbox, (only for 2D networks) when checked, a single model file with labels will be used
- The Labels extension ▼ dropdown, (only for 2D networks) is used to select extension of files containing models. For 3D network MIB model format is used
- The Number of classes edit box, (TIF or PNG formats only) is used to define number of classes (including Exterior) in labels. For label files in MIB *.model format, this field will be updated automatically
- [✓] Use masking checkbox is used when some parts of the training
data should be excluded from training. The masks may be provided in various formats
and number of mask files should match the number of image files. When
mask files are provided the preprocessing operation has to be done. When USE 0-s IN LABELS ▼
is selected the mask is assumed to be areas of label files with 0-values.
This option is recommended for work with masks without the preprocessing operation
- When USE 0-s IN LABELS ▼ is used, the
first material in the prediction results will be assigned to the
Exterior material, i.e. will acquire index 0.
It is recommended to have the first material in the ground truth assigned to background! - When mask with the preprocessing operation is used, the Exterior material will be used to indicate the background areas
- Masking may give drop in precision of training due to inconsistency within the image patches, it is recommended to minimize use of masking
- When USE 0-s IN LABELS ▼ is used, the
first material in the prediction results will be assigned to the
Exterior material, i.e. will acquire index 0.
- Mask extension ▼ is used to select extension for files that contain masks. Without preprocessing (USE 0-s IN LABELS ▼ any files are allowed); with preprocessing only *.mask format is allowed for the 3D network
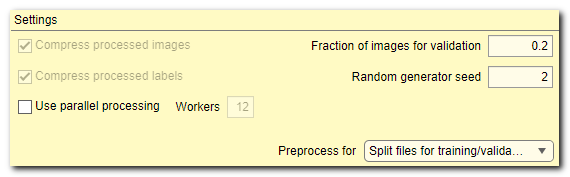
- [✓] Compress processed images, tick to compress the processed images.
The processed images are stored in *.mibImg format that can be loaded in MIB.
*.mibImg is a variation of standard MATLAB format and can also be directly loaded into MATLAB
using similar to this command:
res = load('img01.mibImg, '-mat');.
Compression of images slows down performance! - [✓] Compress processed labels, tick to compress labels during preprocessing.
The processed labels are stored in *.mibCat format that can be loaded in MIB (Menu->Models->Load model).
It is a variation of a standard MATLAB format, where the model is encoded using categorical class of MATLAB.
Compression of labels slows down performance but brings significant benefit of small file sizes - [✓] Use parallel processing, when ticked DeepMIB is using multiple cores to process images. Number of cores can be specified using the Workers edit box. The parallel processing during preprocessing operation brings significant decrease in time required for preprocessing.
- Fraction of images for validation, define fraction of images that will be randomly (depending on Random generator seed) assigned into the validation set. When set to 0, the validation option will not be used during the training
- Random generator seed, a number to initialize random seed generator, which defines how the images for training and validation are split. For reproducibility of tests keep value fixed. When random seed is initialized with 0, the random seed generator is shuffled based on the current system time
- Preprocess for ▼, select mode of operation upon press of the Preprocess button. Results of the preprocessing operation for each mode are presented in schemes below
- when labels are stored in a single *.MODEL file
- when training set is coming in proprietary formats that can only be read using BioFormats reader
- Training and prediction, to preprocess images for both training and prediction
- Training, to preprocess images only for training
- Prediction, to preprocess images only for prediction
- 1_Training\TrainImages, images to be used for training
- 1_Training\ValidationImages, images to be used for validation (optionally)
Additional settings
Preprocessing of files
Originally, the preprocessing of files in DeepMIB was required for most of workflows. Currently, however, DeepMIB is capable to work with unprocessed images most of times: use the Preprocessing is not required ▼ or Split for training/validation ▼ options.
When the preprocessing step is required or recommended
The preprocessing is recommended/required in the following situations:
During preprocessing the images and model files are converted to mibImg and mibCat formats (a variation of MATLAB standard data format) that are adapted for training and prediction.
Organization of directories without preprocessing for semantic segmentation
Without preprocessing, when datasets are manually split into training and validation sets
In this mode, the training image files are not preprocessed and loaded on-demand during network training. The image files should be split into subfolders TrainImages, TrainLabels and optional subfolders ValidationImages, ValidationLabels (for details see Snapshot with the legend below). The images may also be automatically split, see
When Bio-Format library is used, it is recommended to
Snapshot with the directory tree
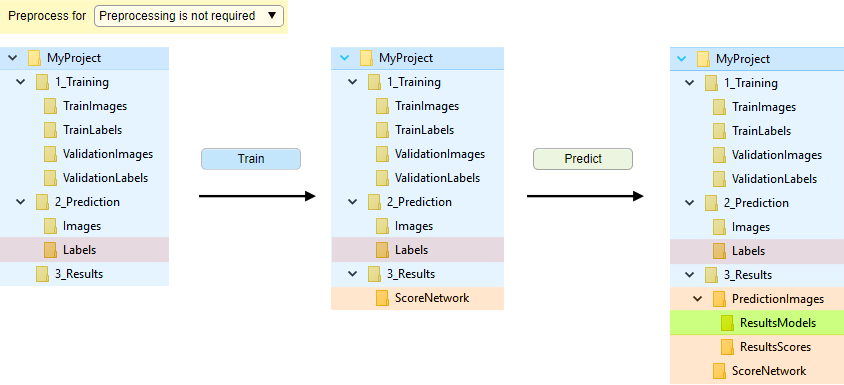
Snapshot with the legend

Without preprocessing, with automatic splitting of datasets into training and validation sets
In this mode, the image and label files are randomly split into the train and validation sets. The split is done upon press of the Preprocess button, when Preprocess for: Split for training and validation
Splitting of the files depends on a seed value provided in the Random generator seed field; when seed is 0 a new random seed value is used each time the spliting is done.
Snapshot with the directory tree
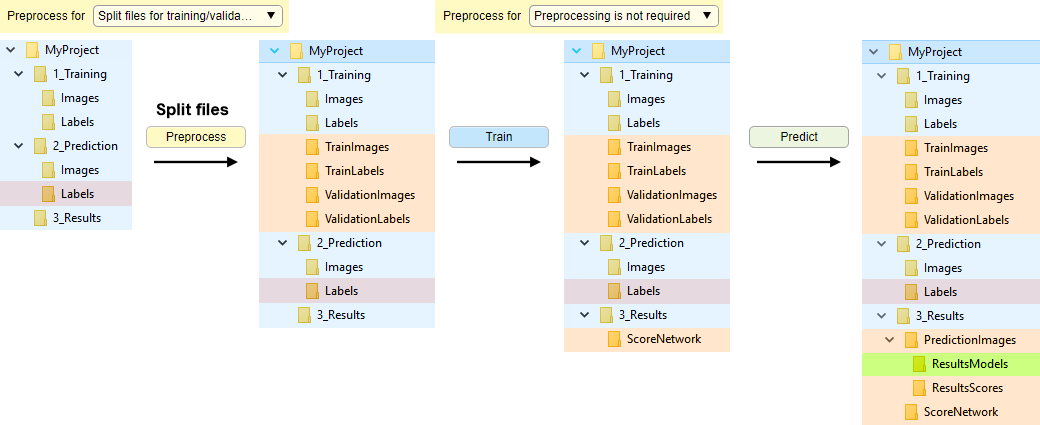
Snapshot with the legend

Organization of directories with preprocessing for semantic segmentation
This mode is enabled when the Preprocess for has one of the following selections:
The preprocessing starts by pressing of the Preprocess button.
The scheme below demonstrates organization of directories, when the preprocessing mode is used.
Snapshot with the directory tree
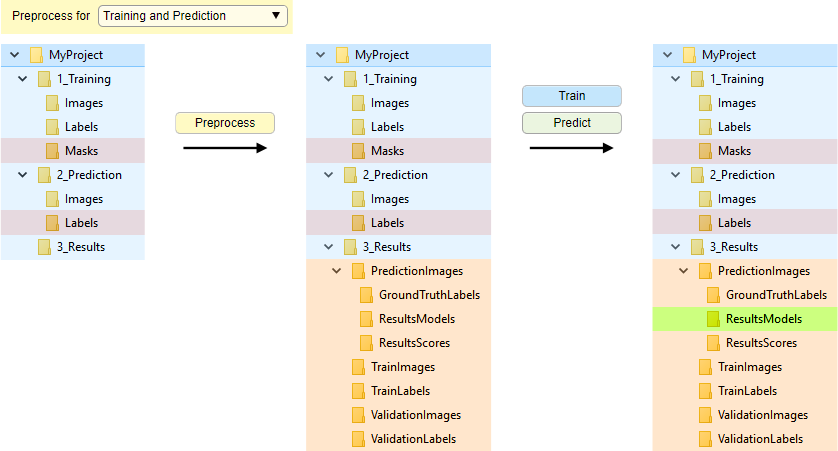
Snapshot with the legend

Organization of directories for 2D patch-wise workflow
The 2D patch-wise workflow requires slightly different organization of images in folders. In brief, instead having Images and Labels training directories, all images are organized in Images\[ClassnameN] subfolders. Where ClassnameN encodes a directory name with images patches that belong to ClassnameN class. Number of these subfolders should match number of classes to be used for training.
In contrast to semantic segmentation, the preprocessing is not used during the patch-wise mode.
Without preprocessing, when datasets are manually split into training and validation sets
The images for training should be organized in own subfolders named by corresponding class names and placed under:
The images may also be
Snapshot with the directory tree
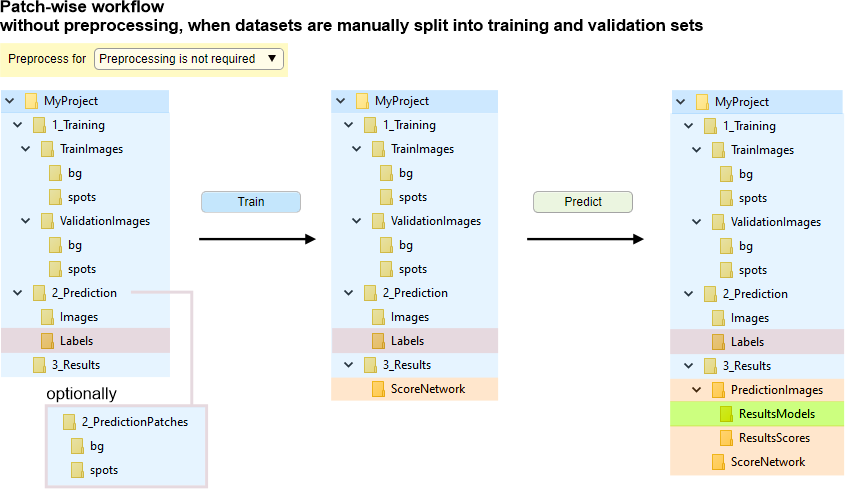
bg and spots are examples of two class names
When the ground-truth data for prediction is present, it can be arranged in a similar way to the semantic segmentation under
2_Prediction\Images and
2_Prediction\Labels directories
or in subfolders named by class names as 2_Prediction\bg and 2_Prediction\spots, where bg and spots subfolders contain patches that belong to these classes.
Snapshot with the legend

Without preprocessing, with automatic splitting of datasets into training and validation sets
In this mode, the files are randomly split (depending on
Initially, all images for training should be organized in own subfolders named by corresponding class names and placed under: 1_Training\Images
Snapshot with the directory tree
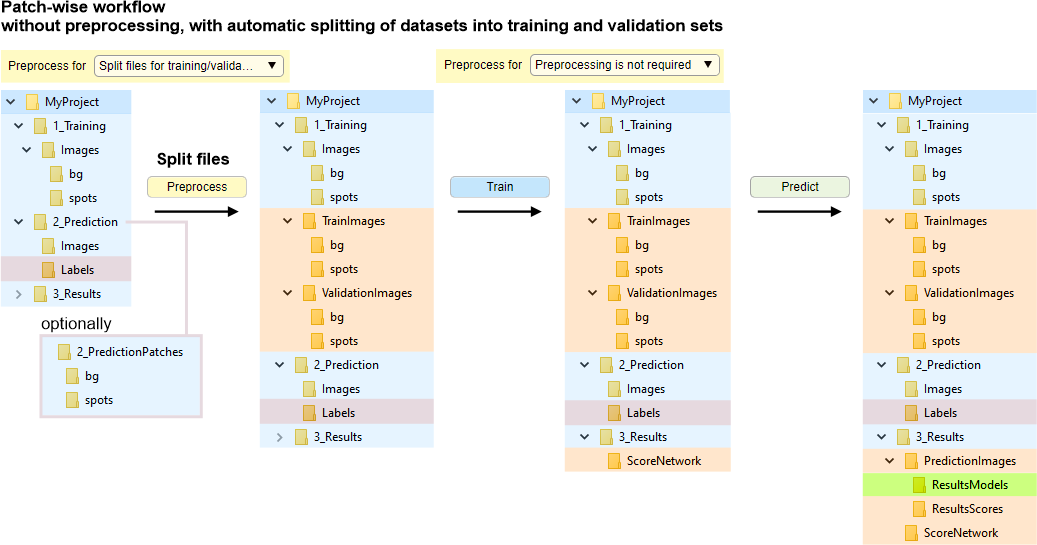
bg and spots are examples of two class names
When the ground-truth data for prediction is present, it can be arranged in a similar way to the semantic segmentation under
2_Prediction\Images and
2_Prediction\Labels directories
or in subfolders named by class names as 2_Prediction\bg and 2_Prediction\spots, where bg and spots subfolders contain patches that belong to these classes.
Snapshot with the legend

Back to Index --> User Guide --> Menu --> Tools Menu --> Deep learning segmentation