Alignment of datasets with Microscopy Image Browser
Ilya Belevich
Dataset: FEI Dual-Beam SEM
Alignment tool can be used to align two adjacent datasets or to align slices within the currently opened dataset.
In addition to the image layer the other layers (selection, mask and overlay) are also aligned.
1. Download and unzip the dataset
2. Open the first part of the dataset in the Microscopy Image Browser
�
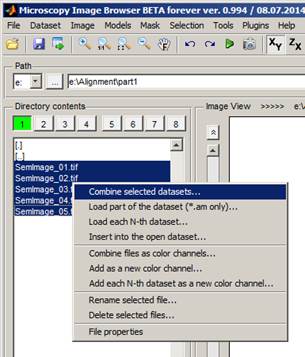 Find
directory with the first part of the dataset in the Directory contents
panel
Find
directory with the first part of the dataset in the Directory contents
panel
� Highlight files in the Directory contents panel. Click with the left mouse button on SemImage_01.tif, hold the Shift key and left mouse click on SemImage_05.tif
� Press the right mouse button to call a popup menu
� Select Combine selected datasets...
3. Start the Alignment tool. Menu->Dataset->Alignment tool...
4. Select Mode->Align two stacks to generate a single stack from two parts of the image dataset.
5. Give path to directory that has the second part of the dataset.
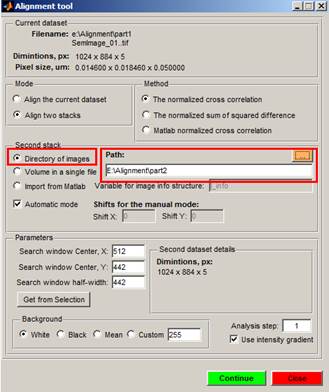
It is also possible to have the second dataset as a single 3D volume file, or import it from the main Matlab workspace.
6. Select one of the Methods (The normalized cross correlation, the normalized sum of squared differences, or Matlab normalized cross correlation) to use for alignment
7. Define parameters, such as Search window, center X, Y and half-width. These values define a search window that will be used to find a point for alignment.
8. Define a color for the background: white, black, mean or provide a custom color.
9. Press the Continue button to align two parts of the dataset. If the alignment procedure fails, it is possible to align datasets manually by providing the X and Y shift values (uncheck the automatic mode checkbox). Or realign them later.
10. Now the both stacks are aligned with each other, however the individual slices are not aligned, please proceed further to align slices.
Align slices in the current dataset
1. Start the Alignment tool again.
2. Select the Align the current dataset mode
3. Press the Continue button to do the alignment.
If the alignment procedure fails, it is possible to select another area for the search window.
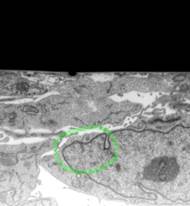
� Draw a circle around area that has good contrast using the Brush tool (Segmentation panel->Selection type->B rush)
� Start the Alignment tool and press Get from Selection button to alter position of the searching window.
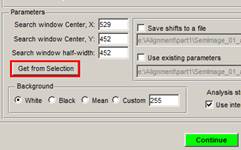
� Press the Continue button.
The X and Y shifts between the slices can be saved (Parameters->Save shifts to a file) and reused again (Parameters->Use existing parameters).
Alignment with landmarks
It is possible to align dataset using landmarks. To do so
1. Select the Brush or Spot tool, Segmentation panel->Selection type->Brush tool
2. Mark corresponding points with a spot on two consecutive slices:
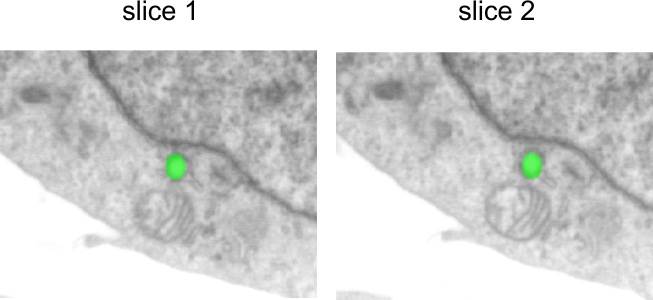
3. Start the Alignment tool
4. Select Options->Landmarks
5. Press the Continue button to align the stack