Overview of Visualization Possibilities with Microscopy Image Browser
Ilya Belevich
Dataset: Huh7.tif (original), obtained by Serial Block Face-SEM (Gatan 3View)
Huh7_crop.tif (cropped)
Dimensions: 32.58 x 32.58 x 2.22 μm (original)
7.01 x 4.99 x 2.22 μm (cropped)
Pixel size: 13.45 x 13.45 x 30 nm
Link to the dataset: http://mib.helsinki.fi/tutorials/3D_Modeling_files/Huh7.tif
Link to the model:
http://mib.helsinki.fi/tutorials/3D_Modeling_files/Model.mat
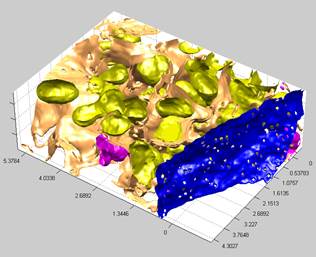
1. Visualization with Matlab:
- Select Segmentation Panel->Materials->Show all to visualize all materials of the model, or uncheck this checkbox and select material that has to be visualized in the list below the checkbox.
- press the right mouse button to call a popup menu,
- select 'Show isosurface (Matlab)'
- in a dialog enter simplification parameters and press OK.
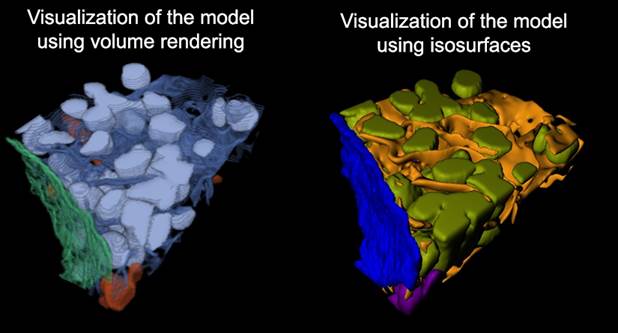
2. Visualization with Fiji. The models can also be
visualized with Fiji/ImageJ 3D viewer (see more here).
In brief:
- Select Segmentation Panel->Materials->Show all to visualize all materials of the model, or uncheck this checkbox and select material that has to be visualized in the list below the checkbox.
- press the right mouse button to call a popup menu,
- select 'Show as Volume (Fiji).'; requires Fiji to be installed.
- in a dialog enter simplification parameter and press OK.
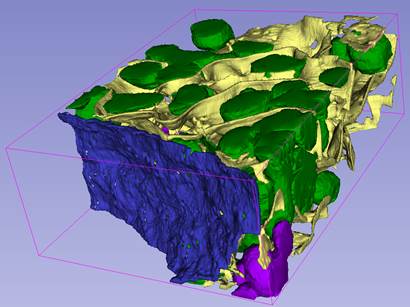
3. Visualization with Slicer 3D.
A video tutorial on youtube.com
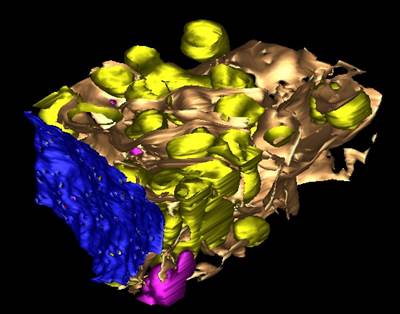
4. Visualization with IMOD.
Please refer to Export to IMOD tutorial for details
5. Visualization with Amira (Visualization Sciences Group, FEI Company).
Shown in brief in the MIB: Image Segmentation tutorial 8: fixing mitochondria and final visualization tutorial.

6. Visualization with Imaris (Bitplane, Oxford Instruments).
Please refer to Render in Imaris (html, pdf, youtube (part 1), youtube (part2)) tutorials for details
