Microscopy Image Browser Watershed/Graphcut segmentation
Tools for separation of objects that can be as materials of the current model, the mask or selection layers.
Back to Index --> User Guide --> Menu --> Tools Menu
Contents
-
General example
-
Mode panel
-
Subarea panel
-
Object separation settings
-
Object separation example
-
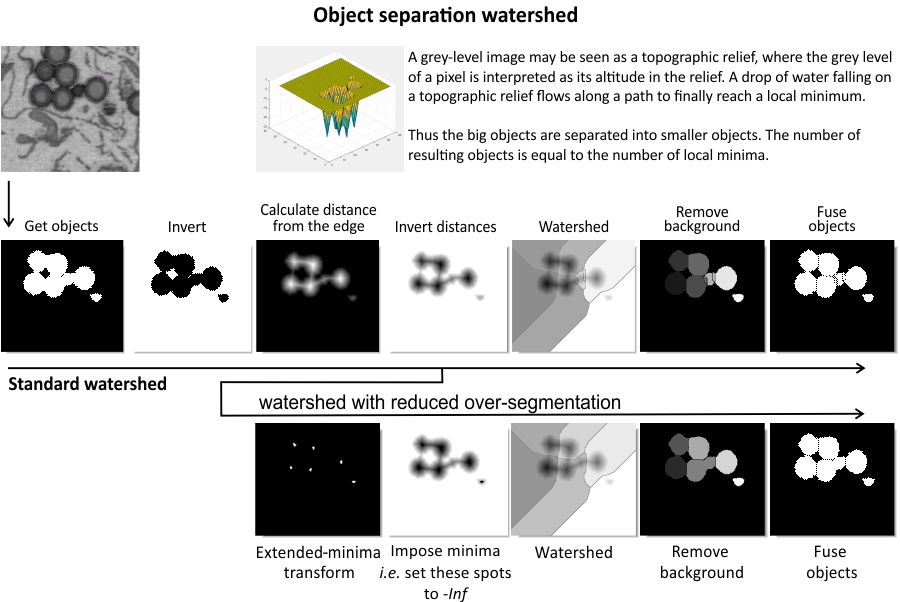
Algorithm for object separation with watershed
-
References
General example
Mode panel
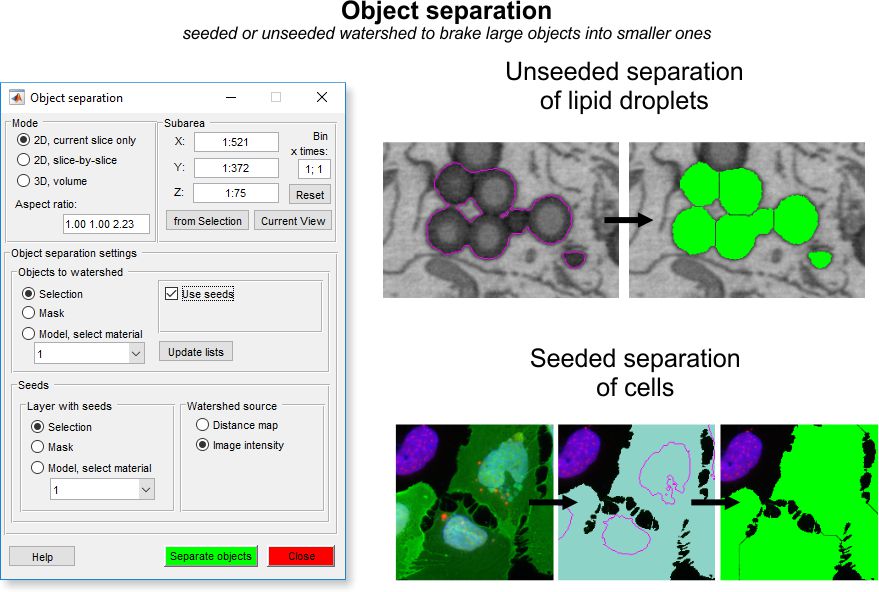
The Mode panel offers possibility to select a desired working mode for the segmentation.
|
- ◉ 2D, current slice only, performs segmentation on the slice that is currently shown
in the Image View panel
- ◉ 2D, slice-by-slice, performs 2D segmentation for each slice of the
dataset individually
- ◉ 3D, volume, performs 3D segmentation for complete or selected portion (see Selected Area section below) of the dataset
- Aspect ratio for 3D... indicates the aspect ratio of the dataset. These values are calculated from the voxel size of the dataset (available from the
Menu->Dataset->Parameters). The aspect ratio
values are used when watershed is running using the distance map (see below)
|
Subarea panel
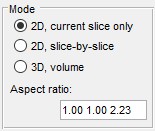
The Subarea panel allows selection of the sub-area of the dataset for processing. If dataset is too big it can be processed in parts or binned using this panel.
|
- X:... defines the width of the dataset to process. Please use two numbers separated by a colon sign (:)
- Y:... defines the height of the dataset to process
- Z:... defines the z-slices of the dataset to process
- from Selection button populates the X:, Y:, Z: fields
using coordinates of a bounding box that describes the Selection layer
- Current View button limits the X: and Y: parameters to the image
that is currently displayed in the Image View panel
- Reset resets the Subarea fields to the dimensions of the dataset
- Bin x times... defines a binning factor for the data before segmentation.
It allows to perform faster but with less details.
Attention! Use of binning during
the Object separation mode may give unpredictable results
|
Object separation settings
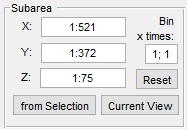
The Object separation mode uses the watershed transformation to brake segmented objects into smaller ones. The specific settings for the Object separation mode are shown below.
|
- Object to watershed defines a layer that contains a source object for separation. It could be one of the main layers: Selection, Mask, or Model
- [✓] Use seeds when enabled targets algorithm to the seeded watershed transformation. Some parameters should be additionally specified in the Seeds panel
- Reduce oversegmentaion (available only for the unseeded watershed transformation) decreases number of resulting objects
- Seeds panel (only for the seeded watershed transformation)
- Layer with seeds defines a layer that contains seeds. It could be one of the main layers: Selection, Mask, or Model
- Watershed source defines type of the information that watershed will use for labeling.
When the Image intensity option is selected watershed is using actual image intensities rather than the distance maps.
See more in the Steve Eddins's blog on Image Processing.
|
Object separation example
The object separation with watershed can be used to separate big objects into smaller ones. For example, some mitochondria from the image segmentation example are fused together. It is possible to separate them using the Object separation mode.
-
Menu->Tools->Object separation
- Select Mask in the Objects to watershed
- Press the Segment button
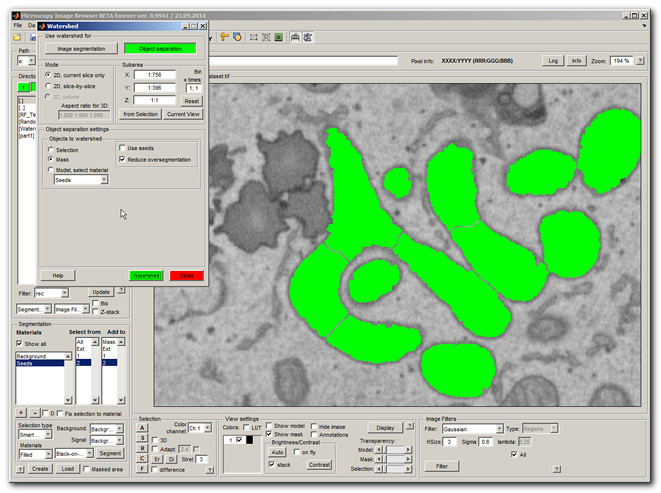
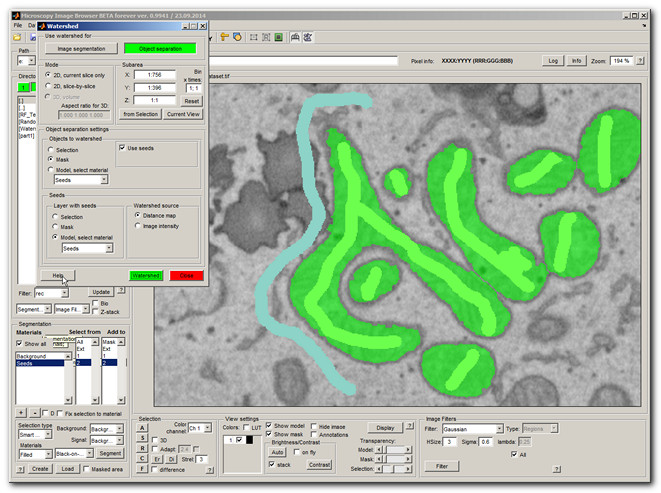
Now the mitochondria are separated, but unfortunately, as usual with watershed, long mitochondria are broken into several small pieces as well. To deal with that the seeded watershed can be used.
- Press the [✓] Use seeds checkbox
- Choose Model, select material -> 'Seeds' in the Layer with seeds panel
- Press the Segment button
- If there is only one label in each mitochondria the individual mutochondria should be extracted (shown in green)
Algorithm for object separation with watershed
References
Watershed: the Image segmentation and Object separation modes:
Back to Index --> User Guide --> Menu --> Tools Menu






