

Navigation
System Requirements
Contents:
|
Computer MATLAB Toolboxes |
|
| -------------------- Optional -------------------- | |
|
Bio-Formats BMxD filters DipLib library Edge Enhancing Coherence Filter Fiji Frangi Filter Imaris |
Membrane Click Tracker Omero Server Random Forest Classifier Read NRRD Segment-anything model Supervoxels/Graphcut Volume Rendering Zarr format |
It is recommended to install SAM2 as it gives faster and better results.
If needed, SAM-1 can be added on a top of SAM-2 installation.
If needed, SAM-1 can be added on a top of SAM-2 installation.
Contents
General information
Segment-anything model (SAM) is developed by Meta AI Research, FAIR
research team. It can be used to segment individual objects or the whole image using a single mouse click.
Details of the research is available here: https://segment-anything.comImplementation of SAM in MIB is done via utilization of an external Python interpreter, please follow the following steps to configure local python environment to work in MIB.
Important! even though SAM can work on CPU, GPU is highly recommended as it is x30-60 faster.
Reference
-
Alexander Kirillov, Eric Mintun, Nikhila Ravi, Hanzi Mao, Chloe Rolland, Laura Gustafson, Tete Xiao,
Spencer Whitehead, Alexander C. Berg, Wan-Yen Lo, Piotr Dollar, Ross Girshick
Segment Anything
arXiv:2304.02643, https://doi.org/10.48550/arXiv.2304.02643
Requirements
- MATLAB R2022a or newer (tested on R2022a, R2022b, R2023a)
- Python 3.8, 3.9, 3.10; tested on 3.9
- List of Python versions compatible with various MATLAB releases
- CUDA-compatible GPU is highly recommended, CPU can also be used but it is significantly slower
Installation on Linux
SAM can also be used under Linux, for details please check the dedicated page:https://mib.helsinki.fi/downloads_systemreq_sam_linux.html
------------- Installation instructions -------------
Installation and usage:
Python installation
- Install Miniconda
(tested on python 3.9, version 23.1.0, Miniconda3-py39_23.1.0-1-Windows-x86_64.exe)
https://docs.conda.io/en/latest/miniconda.html
Archive of miniconda releases: https://repo.anaconda.com/miniconda/
If you have admin account, you can install Python for all users, otherwise it is possible to install Python only for the current user.
- With Admin rights
Example of the installation directory:D:\Python\Miniconda39\


Optional note!
If installation was done for All Users, change permission of Miniconda'senvsdirectory (e.g.d:\Python\Miniconda39\envs\) or the whole Miniconda39 directory to be accessible for all users.
This makes things a bit more organized, otherwise the python environment will be created inC:\Users\[USERNAME]\.conda\envs\


- Without Admin rights
Example of the installation directory:C:\Users\[USERNAME]\AppData\Local\miniconda39\



Install segment-anything model
- Download segment-anything from:
- forked distibution tested with MIB
- facebookresearch github

- Unzip "segment-anything-main.zip", for example to
d:\Python\Examples\segment-anything\
Install required python packages
- Create a new environment for Python with SAM
- Start "Anaconda Prompt"
Start->Miniconda3->Anaconda prompt (Miniconda39)

- Create a new environment, specify location for the environment and the version of Python:
>> conda create --prefix d:\Python\Miniconda39\envs\sam4mib python=3.9
- Start "Anaconda Prompt"
- Activate the environment:
>> activate sam4mib - The code requires python>=3.8, as well as pytorch>=1.7 and torchvision>=0.8.
the instructions here to install both PyTorch and TorchVision dependencies:
https://pytorch.org/get-started/locallyThe forked distribution has"requirements.txt"that can be used to install required dependencies at once for Windows with CUDA 11.8 supported GPU:
- Make sure the the environment is created and activated (steps above)
- In the terminal/anaconda prompt run the command to install required packages
(make sure that the correct path to requirements.txt is specified):
>> pip3 install -r requirements.txt
or
>> pip3 install -r d:\Python\Examples\segment-anything\requirements.txt - Proceed to the MIB configuration section below
- The following dependencies are necessary for mask post-processing, saving masks in COCO format,
the example notebooks, and exporting the model in ONNX format:
>> pip3 install opencv-python matplotlib onnxruntime onnx
>> pip3 install pycocotools
if there is an error see below - Install "onnxruntime-gpu" to make prediction on GPU:
>> pip3 install onnxruntime-gpu==1.14.1 - Optionally, install Jupyter notebook:
>> pip install notebook
MIB configuration
- Start MIB
- Open MIB preferences:
Menu->File->Preferences - Define path of
python.exeinstalled in the specified environment (sam4mib):
External directories->Python installation path
For example:
-D:\Python\Miniconda39\envs\sam4mib\python.exe
-C:\Users\[USERNAME]\.conda\envs\sam4mib\python.exe
- Define directory to store network architectures for DeepMIB;
this location will be used to download checkpoints and onnx models.
- Select "Segment-anything model" tool in the Segmentation panel:
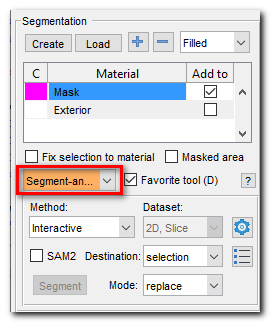
- Open SAM settings:
Segmentation panel->Segment-anything model->Settings
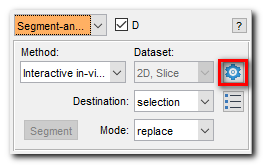

- Select the backbone:
-vit_b (0.4Gb), fastest (x1) but gives less precise results
-vit_l (1.2Gb), moderate speed (~x1.4 slower), better predictions
-vit_h (2.5Gb), slowest (x2.0), best predictions

- Define location where segment-anything package was unzipped:
if you checkCheck to select path to segment-anythinga directory selection dialog will be shown

- Set correct execution environment, please note that CPU is 30-60 times slower than CUDA

How to use
Documentation on segment-anything is available:
- youtube tutorial (SAM1 version):

- Documentation page is available in MIB Help:
User guide->
...GUI Overview, Panels->
......Segmentation panel->
..........Segmentation tools->Segment-anything model
JSON file for custom networks
All trained SAM networks are automatically installed upon user's demand. Links to the these networks are specified in
sam_links.json file located in Resources subfolder in MIB installation directory.This json file can be used to specify locations to custom networks and those networks will be automatically linked and become available under the backbone section of the SAM configuration window.
An example of a section within json file that specify location of "Standard SAM vit_h model"; use this as a template if you need to add a custom trained network
{
"info": "Standard SAM vit_h model",
"name": "vit_h (2.5Gb)",
"backbone": "vit_h",
"checkpointFilename": "sam_vit_h_4b8939.pth",
"onnxFilename": "sam_vit_h_4b8939_quantized.onnx",
"checkpointLink_url_1": "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_h_4b8939.pth",
"checkpointLink_url_2": "http://mib.helsinki.fi/web-update/sam/sam_vit_h_4b8939.pth",
"onnxLink": "http://mib.helsinki.fi/web-update/sam/vit_h.zip"
}
Description of fields:- info: information about the SAM trained network
- name: name how it will be shown in the SAM configuration
- backbone: backbone name
- checkpointFilename: filename of the network file
- onnxFilename: [only for SAM1], onnx filename that is used with SAM version 1, not used for SAM2
- checkpointLink_url_1: an URL link from where pytorch file will be downloaded
- checkpointLink_url_2: an alternative URL link, in case when the first one does not work
- modelCfgLink_url_1: [only for SAM2] an URL link to the config file, that is typically located under sam2_configs directrory of SAM2
- modelCfgLink_url_2: [only for SAM2] alternative link to the config file
- onnxLink: [only for SAM1] an URL to a generated onnx file
Troubleshooting
error: Microsoft Visual C++ 14.0 or greater is requiredThis error may appear during
Solution 1:
- Try to solve the issue by installing the required package via conda using this command:
>> conda install -c conda-forge pycocotools
Solution 2:
- Navigate to https://visualstudio.microsoft.com/visual-cpp-build-tools/
- Download Build Tools

- Install "Desktop development with C++"

- Type the command again
>> pip install opencv-python pycocotools matplotlib onnxruntime onnx
Remove sam4mib environment
If you do not need mib4sam environment, you can follow the following steps to uninstall it from your system.
- Start "Anaconda Prompt"
Start->Miniconda3->Anaconda prompt (Miniconda39)

- List the environments installed on your system by running the command:
>> conda env list - Remove sam4mib environment:
>> conda remove --name sam4mib --all

