Microscopy Image Browser Watershed/Graphcut segmentation
This window give access to semi-automated image segmentation using the Watershed method.
It is recommended to use the Graphcut segmentation due to its high interactivity.
Back to Index --> User Guide --> Menu --> Tools Menu
Contents
-
General example
-
Mode panel
-
Subarea panel
-
Image segmentation settings
-
Image segmentation example
-
Algorithm for image segmentation with watershed
-
References
General example
Mode panel
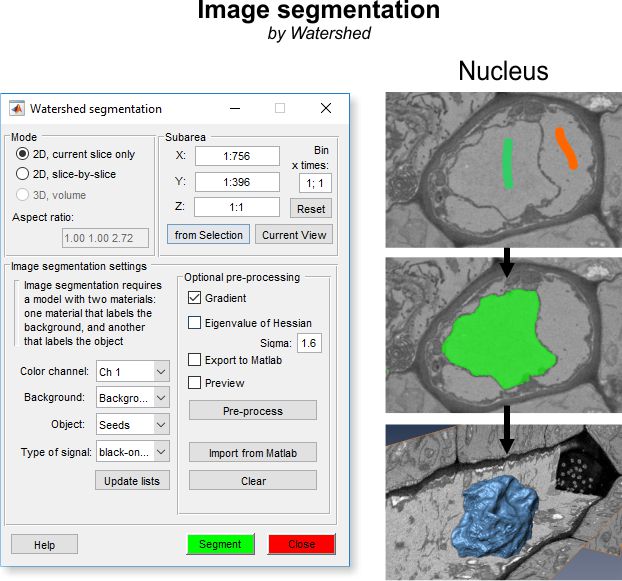
The Mode panel offers possibility to select a desired working mode for the segmentation.
|
- ◉ 2D, current slice only, performs segmentation on the slice that is currently shown
in the Image View panel
- ◉ 2D, slice-by-slice, performs 2D segmentation for each slice of the
dataset individually
- ◉ 3D, volume, performs 3D segmentation for complete or selected portion (see Selected Area section below) of the dataset
- Aspect ratio for 3D... indicates the aspect ratio of the dataset.
These values are calculated from the voxel size of the dataset (available from the
Menu->Dataset->Parameters). The aspect ratio
values are used when watershed is running using the distance map (see below)
|
Subarea panel
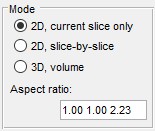
The Subarea panel allows selection of the sub-area of the dataset for processing. If dataset is too big it can be processed in parts or binned using this panel.
|
- X:... defines the width of the dataset to process. Please use two numbers separated by a colon sign (:)
- Y:... defines the height of the dataset to process
- Z:... defines the z-slices of the dataset to process
- from Selection button populates the X:, Y:, Z: fields
using coordinates of a bounding box that describes the Selection layer
- Current View button limits the X: and Y: parameters to the image
that is currently displayed in the Image View panel
- Reset resets the Subarea fields to the dimensions of the dataset
- Bin x times... defines a binning factor for the data before segmentation.
It allows to perform faster but with less
details.
|
Image segmentation settings
Both the Watershed and Graphcut segmentation workflows use provided labels that mark areas belonging to the Object and Background to perform the fine segmentation. Comparing to the Graphcut workflow, the Watershed workflow is a bit less interactive; it requires more time for the each execution and separates only objects that have distinct boundaries, for example membrane enclosed organelles.
On the other hand, the Graphcut workflow spends more time on the image preprocessing (calculation of the superpixels and generation of a graph) but each following interaction is fast. Using this workflow it is possible to separate objects that have both boundaries and intensity contrast. In general the Graphcut workflow is recommended for most of the cases.
Below, description of the Image segmentation settings:
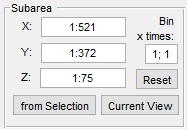
|
- Color channel ▼ defines a color channel that will be used for
segmentation
- Background ▼ defines a material of the model that labels the
background
- Object ▼ defines a material of the model that labels the
object to be segmented
- Type of signal ▼ defines type of the data: 'black-on-white', when the
objects are separated with dark boundaries and 'white-on-black' for the
bright boundaries
- Update lists refreshes the lists of materials
- Optional pre-processing (only for the Watershed workflow)
- [✓] Gradient filters the image before watershed using the
Gradient filter to create borders around objects
- [✓] Eigenvalue of Hessian, pre-processing the data using this option may sometimes be beneficial for the following watershed transfornation. Use the Sigma fields to fine-tune the filter
- [✓] Export to MATLAB exports pre-processed data to the main MATLAB workspace
- [✓] Preview shows the result of pre-processing in the Image View panel
- Pre-process starts the data pre-processing process. When pre-processed data is present the color of the button turns to green
- Import from MATLAB imports dataset that will be used for image segmentation from MATLAB workspace
- Clear removes the pre-processed data from the memory
|
Image segmentation example
|
- Load a sample dataset: Menu->File->Import image from->URL, enter the address:
http://mib.helsinki.fi/tutorials/WatershedDemo/watershed_demo1.tif
- Press the + button in the Segmentation panel to add material to the model and name is as 'Background' (use the right mouse button to call a popup menu)
- Use the brush tool to label an area that belongs to cytoplasm
|
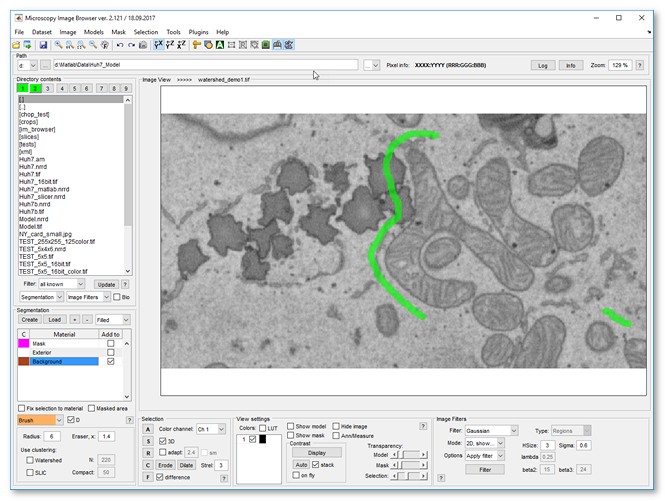
|
- Press the A button to add selected area to the first material (Background) of the model
- Press the + button again to add another material and name it as 'Seeds'
- Draw labels inside mitochondria.
|
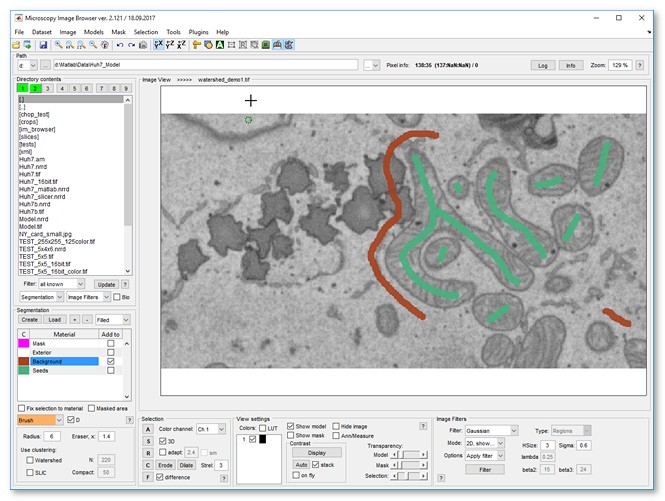
|
- Press the A button to add selected area to the second material (Seeds) of the model
- Start the Watershed segmentation tool: Menu->Tools->Semi-automatic segmentation->Watershed.
- Make sure that the proper materials are selected for both Background and Object in the Image segmentation settings
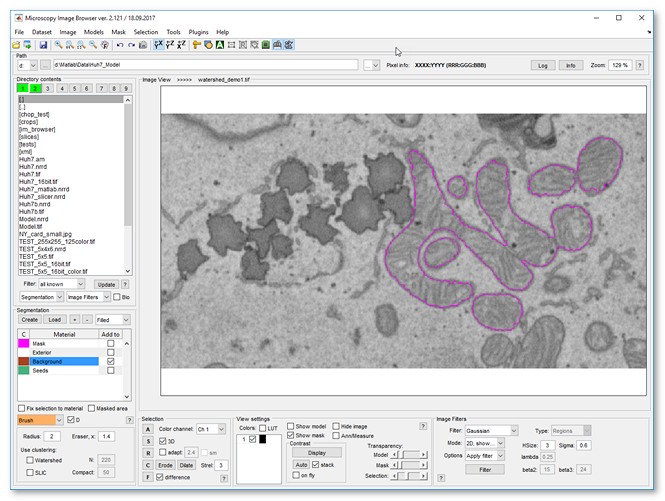
- Press the Segment button to segment mitochondria
- Add more seeds to the background and object materials to improve segmentaion
- Press the Segment button again
- The segmented mitochondria are placed to the Mask layer
- Optionally smooth mitochondria: Menu->Mask->Smooth Mask
|
|
Algorithm for image segmentation with watershed
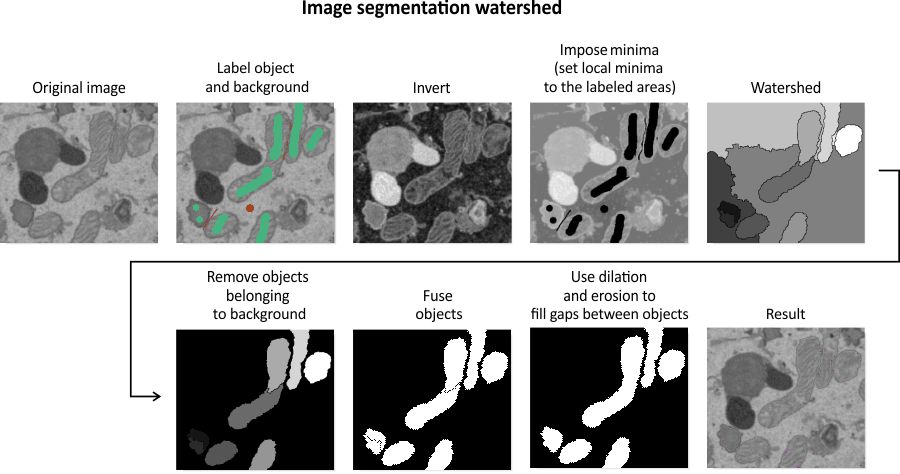
References
Watershed: the Image segmentation and Object separation modes:
Back to Index --> User Guide --> Menu --> Tools Menu








