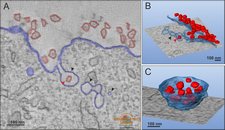
Modeling og plasma membrane internalization of Semliki Forest virus.
Modeling of plasma membrane internalization of Semliki Forest virus.
|
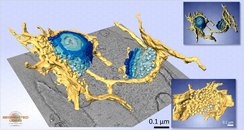
A model of Golgi apparatus and ER exit sites from a ET dataset of Trypanosoma brucei. Upper inset: the rendering with Fiji;
lower inset: ER exit site. Dimensions: 1.77 x 2.10 x 0.49 Ám, voxel size: 1.94 x 1.94 x 1.94 nm. All models except the upper inset were visualized with Amira.
A model of Golgi apparatus and ER exit sites from a ET dataset of Trypanosoma brucei.
|

Two successive 250-nm sections from Huh-7 cells were prepared using high-pressure freezing and freeze substitution and subjected to electron tomography. From the resulting tomogram, microtubules, actin fi laments, and the ER network comprising tubules and highly fenestrated sheets were modeled (blue, red, and yellow, respectively)
Microtubules and actin fi laments can be found in close proximity to the endoplasmic reticulum.
Joensuu M, Belevich I, Rämö O, Nevzorov I, Vihinen H, Puhka M, Witkos TM, Lowe M, Vartiainen MK,
Jokitalo E. Mol Biol Cell. 2014 Apr;25(7):1111-26.
|
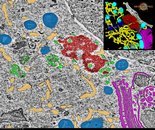
Cytoplasmic surroundings of the chromatoid body (CB). Representative snapshot with a model and the 3D rendering in Fiji (inset) of the segmented structures. The CB (red) was found close to the nuclear membrane (white). Holes visible in the nuclear membrane represent nuclear pores. The CB was surrounded by small cytoplasmic vesicles (green), multivesicular bodies (cyan), mitochondria (blue) and the Golgi complex (purple) were found nearby the CB.
Cytoplasmic surroundings of the chromatoid body.
Da Ros M, Hirvonen N, Olotu O, Toppari J, Kotaja N Mol Cell Endocrinol. 2015 Feb 5;401:73-83. doi: 10.1016/j.mce.2014.11.026
|


 Modeling of plasma membrane internalization of Semliki Forest virus.
Modeling of plasma membrane internalization of Semliki Forest virus.
 A model of Golgi apparatus and ER exit sites from a ET dataset of Trypanosoma brucei.
A model of Golgi apparatus and ER exit sites from a ET dataset of Trypanosoma brucei.
 Microtubules and actin fi laments can be found in close proximity to the endoplasmic reticulum.
Microtubules and actin fi laments can be found in close proximity to the endoplasmic reticulum.
 Cytoplasmic surroundings of the chromatoid body.
Cytoplasmic surroundings of the chromatoid body.
