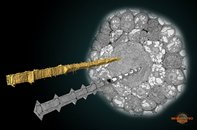
Phloem sieve element cells of Arabidopsis thaliana. The image shows a cross-section of the root and two sieve elements. One of the sieve element is visualized using Amira orthoslices, while another one was modeled in MIB and is shown as a surface. The image was rendered in Amira.
Phloem sieve element cells of Arabidopsis thaliana.
Furuta KM, Yadav SR, Lehesranta S, Belevich I, Miyashima S, Heo JO, Vaten A, Lindgren O, De Rybel B, Van Isterdael G, Somervuo P, Lichtenberger R, Rocha R, Thitamadee S, Tähtiharju S, Auvinen P, Beeckman T, Jokitalo E, Helariutta Y. Science. 2014 Aug 22;345(6199):933-7.
|
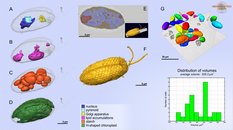
A model of organelles and cell outline of Rhinomonas nottbecki n. sp.
A.-D. models of organelles;
E. an overlay of the segmentation model and the raw slice from 3D dataset; F.a model of the cell surface.
G. A SB-EM volume containing numerous cells and analysis of their volumes.
The models were rendered in Amira.
Cell surface and organelles of Rhinomonas nottbecki n. sp.
Majaneva M, Remonen I, Rintala JM, Belevich I, Kremp A, Setälä O, Jokitalo E, Blomster J.J Eukaryot Microbiol. 2014 Sep-Oct; 61(5):480-92.
|
|
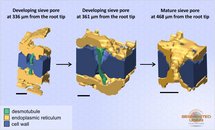
Developing and mature sieve pores in wild-type Col-0 Arabidopsis thaliana. Scale bar: 0.2 µm. The model was rendered in Amira.
Developing and mature sieve pores in wild-type Col-0 Arabidopsis thaliana.
Dettmer J, Ursache R, Campilho A, Miyashima S, Belevich I, O'Regan S, Mullendore DL, Yadav SR, Lanz C, Beverina L, Papagni A, Schneeberger K, Weigel D, Stierhof YD, Moritz T, Knoblauch M, Jokitalo E, Helariutta Y. Nat Commun. 2014 Jul 10;5:4276.
|
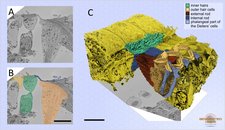
Mouse cochlea was perilymphatically fixed and the sensory epithelium of the medial part of the cochlear duct was imaged. Different cell types of the organ of Corti were segmented using local thresholding and the shape interpolation technique. Scale bar: 10 µm. A. raw data; B. model overlay; C. visualization of the model in Amira.
Modeling of different cell types of the organ of Corti.
Watch video
Anttonen T, Kirjavainen A, Belevich I, Laos M, Richardson WD, Jokitalo E, Brakebusch C, Pirvola U. Sci Rep. 2012;2:978.
Anttonen T, Belevich I, Kirjavainen A, Laos M, Brakebusch C, Jokitalo E, Pirvola U. J Assoc Res Otolaryngol. 2014 Dec;15(6):975-92.
|
|
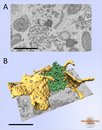
Huh-7 cells expressing ER marker were cytochemically stained and imaged.Model depicting ER sheets (yellow) and tight network of smooth ER tubules (green). Images collected at 30,000x, voxel size 4.7x4.7x25 nm. The model was rendered in Amira. Scale bar: 1 µm. A. raw data; B. visualization of the model.
Modeling of the smooth endoplasmic reticulum.
Watch video
Vihinen H, Belevich I, Jokitalo E. Microscopy and Analysis 27(2) March 2013
Smooth Endoplasmic Reticulum
|
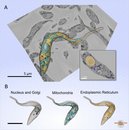
SB-EM dataset of Trypanosoma brucei with one tripanosome segmented from the 3D volume. Dimensions: 11.3 x 15.4 x 6.8 Ám, voxel size: 14 x 14 x 30 nm. A. A raw dataset overlayed with the 3D model; B. Visualization of individual organelles from the model in the panel A. The model was rendered in Amira.
Modeling of different organelles of Trypanosoma brucei.
|
|
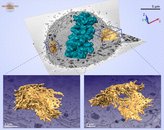
A model of a Huh-7 cell overlaid on one block-face image shows the shape of an early-metaphase cell.
The ER models (insets) show heavily fenestrated sheet structures; however, a more spread network organization was found in the flat part of the cell (right inset),
whereas on the more rounded part of the cell the ER was packed into concentric layers (left inset). The ER membranes are depicted in yellow and the chromosomes in dark cyan.
The model was rendered in Amira.
Models of ER in an early-metaphase Huh-7 cell demonstrating dissimilar ER network organizations within the same cell.
Watch video
Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E. Mol Biol Cell. 2012 Jul;23(13):2424-32.
|
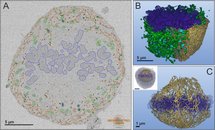
Semi-automatic segmentation of a mitotic cell in the early metaphase.
A. A section of the original dataset of human hepatoma (Huh-7) cells with overlaid modeled organelles. Chromosomes (blue) are concentrated at the center of the cell forming a disk-type structure. Endoplasmic reticulum (ER) forms a shell structure that composed of stacked perforated sheets. In addition to perforated sheets the ER network has extensive tubular components that penetrate into the center of the cell towards chromosomes and correlates with direction of the spindle fibres. Mitochondria are in green and lysosomes are in cyan.
B. A 3D model of a half of the mitotic cell, only part of ER is shown.
C. A 3D model of the tubular ER network that is concentrated around chromosomes. The inset shows relative position of the modeled area within the whole cell. The model was rendered in Amira.
Semi-automatic segmentation of a mitotic cell in the early metaphase.
Watch video
Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E. Mol Biol Cell. 2012 Jul;23(13):2424-32.
|
|



