|
Microscopy Image Browser
1.233
|
 |
Microscopy Image Browser
1.233
|
Get metadata for images. More...
Functions | |
| function [
img_info , files , pixSize ] = | getImageMetadata (filenames, options) |
| Get metadata for images. More... | |
Get metadata for images.
| function [ img_info , files , pixSize ] = getImageMetadata | ( | filenames, | |
| options | |||
| ) |
Get metadata for images.
| filenames | a cell array with filenames of images |
| options | a structure with additional parameters .bioformatsCheck -> 0 or 1; when 1 use the BioFormats library .waitbar -> 0 or 1; when 1 shows the progress bar .customSections -> 0 or 1; when 1 obtain part of the dataset .matlabVersion -> [optional] current version of Matlab .Font -> [optional] a structure with the font settings from im_browser .FontName .FontUnits .FontSize |
| img_info | in the format compatible with imageData.img_info containers.Map |
| files | a structure with file-info, can be empty: []
|
| pixSize | a structure with voxel dimensions (.x, .y, .z, .units, .t, .tunits) |
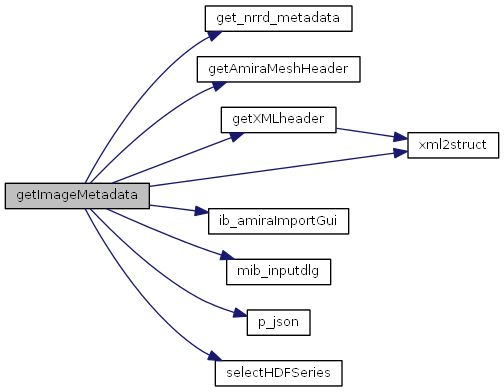
References get_nrrd_metadata(), getAmiraMeshHeader(), getXMLheader(), ib_amiraImportGui(), mib_inputdlg(), p_json(), selectHDFSeries(), and xml2struct().
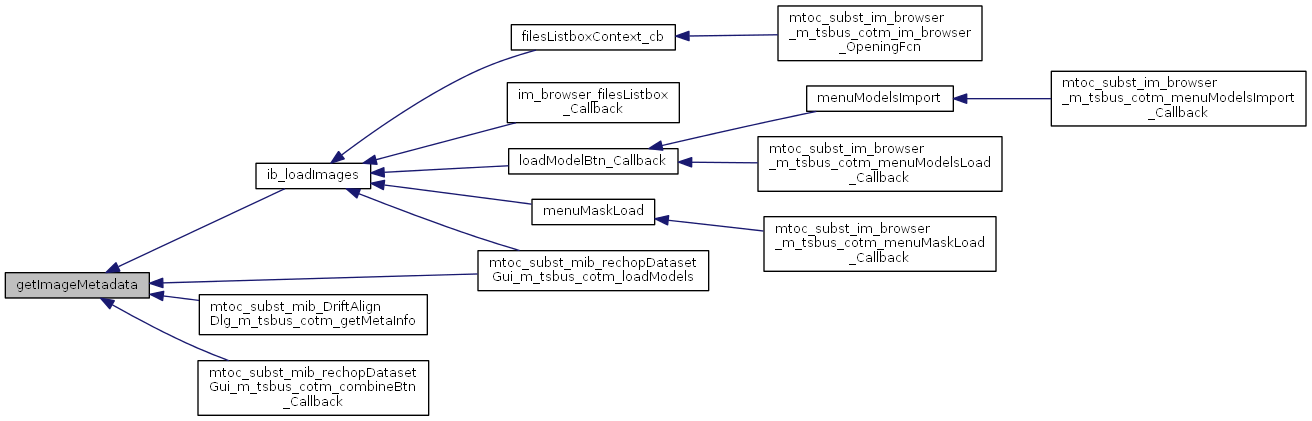
Referenced by ib_loadImages(), mibDriftAlignDlg>getMetaInfo(), mibrechopDatasetGui>combineBtn_Callback(), and mibrechopDatasetGui>loadModels().