Deep MIB - Network Panel
Back to MIB | User interface | DeepMIB
Configuration of workflows and network architectures for deep learning segmentation in Microscopy Image Browser.
Overview
The Network panel in Deep MIB occupies the upper part of the interface and is used to select the workflow and convolutional network architecture for training deep learning models.
Workflows
Start a new project by selecting a workflow:
- 2D Semantic: clusters 2D image pixels of the same material together
- 2.5D Semantic: uses 2D network architectures to process small subvolumes (3-9 stacks), segmenting only the central slice
- 3D Semantic: clusters 3D image voxels of the same material together
- 2D Patch-wise: predicts 2D images in blocks (patches), producing a downsampled image indicating object positions
2D Semantic workflow
list of available network architectures for 2D semantic segmentation
The following architectures are available for 2D semantic segmentation:
-
2D U-net: a convolutional neural network developed for biomedical image segmentation at the University of Freiburg, Germany. it segments a 512x512 image in under a second on a modern GPU (Wikipedia).
References:- Ronneberger, O., et al. "U-Net: Convolutional Networks for Biomedical Image Segmentation." MICCAI, 2015 (arXiv).
- MATLAB U-Net layers.
-
2D SegNet: a convolutional network from the University of Cambridge, UK, designed for general image segmentation, less suited for microscopy data compared to U-net.
References:- Badrinarayanan, V., et al. "SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation." arXiv, 2015 (arXiv).
- MATLAB SegNet layers.
-
2D DeepLabV3 (recommended): an efficient DeepLab v3+ network with selectable base networks (Resnet18, Resnet50, Xception, Inception-ResNet-v2), suitable for diverse segmentation tasks with grayscale or RGB inputs.
- Resnet18: 18-layer network, lightest option, quickest with low GPU needs, 224x224 input size, initialized with a pretrained EM/pathology template (IEEE).
- Resnet50: 50-layer network, balanced performance, 224x224 input size, initialized with a pretrained EM/pathology template (IEEE).
- Xception (MATLAB version only): 71-layer network, 299x299 input size (arXiv).
- Inception-ResNet-v2 (MATLAB version only): 164-layer network, high GPU demands, 299x299 input size (AAAI).
Reference: - Chen, L., et al. "Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation." ECCV, 2018 (arXiv).
2.5D Semantic workflow
Architectures available in 2.5D workflows
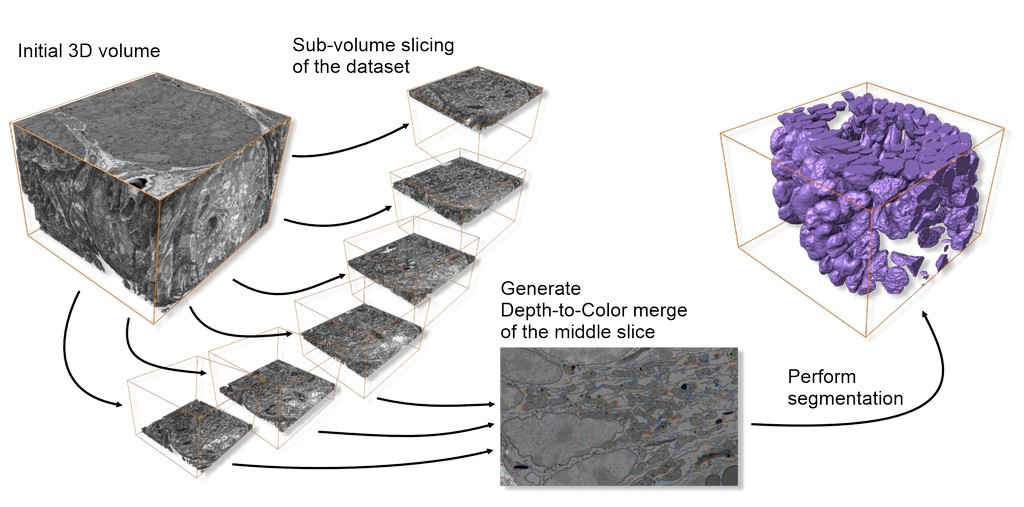
Comparison of 2.5D vs. 2D results:
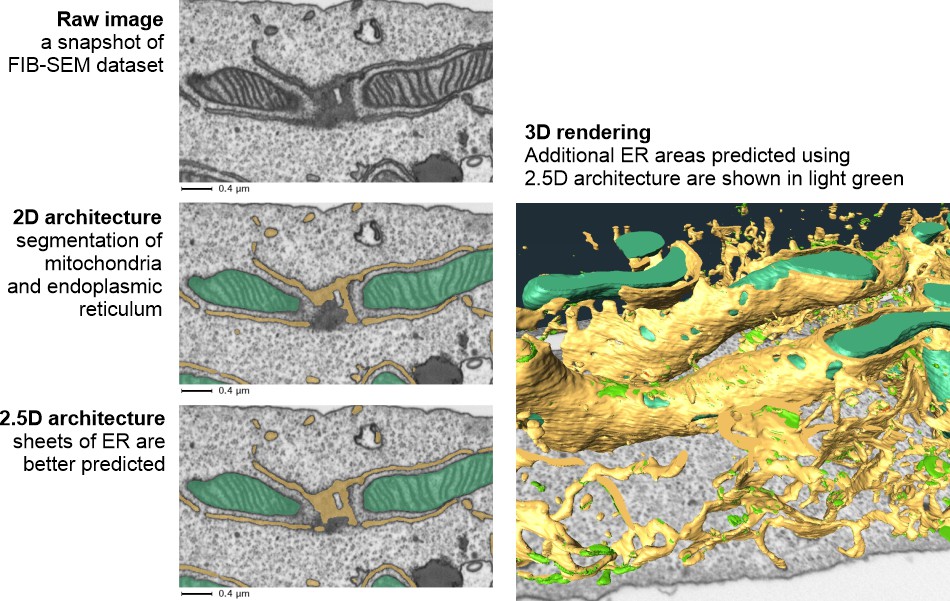
Available architectures (see 2D semantic section for details):
- Z2C + DLv3: DeepLab v3 with Resnet18 or Resnet50 encoders
- Z2C + U-net: standard U-net as the template
- Z2C + U-net + Encoder: U-net with Resnet18 or Resnet50 encoders
Notes on application of 2.5D workflows
Work with 2.5D workflows mirrors 2D workflows, with these differences:
- Input images and labels: use small substacks (3-9 sections), segmenting only the central slice, saved as separate files
- Generation of patches for training: auto-generate subvolumes via:
- From models:
Menu → Models (or Masks) → Model (Mask) statistics → detect objects → right-click → Crop to a file
- From annotations:
Menu → Models → Annotations → right-click over selected annotations → Crop out patches around selected annotations
See Generation of patches for deep learning segmentation
3D Semantic workflow
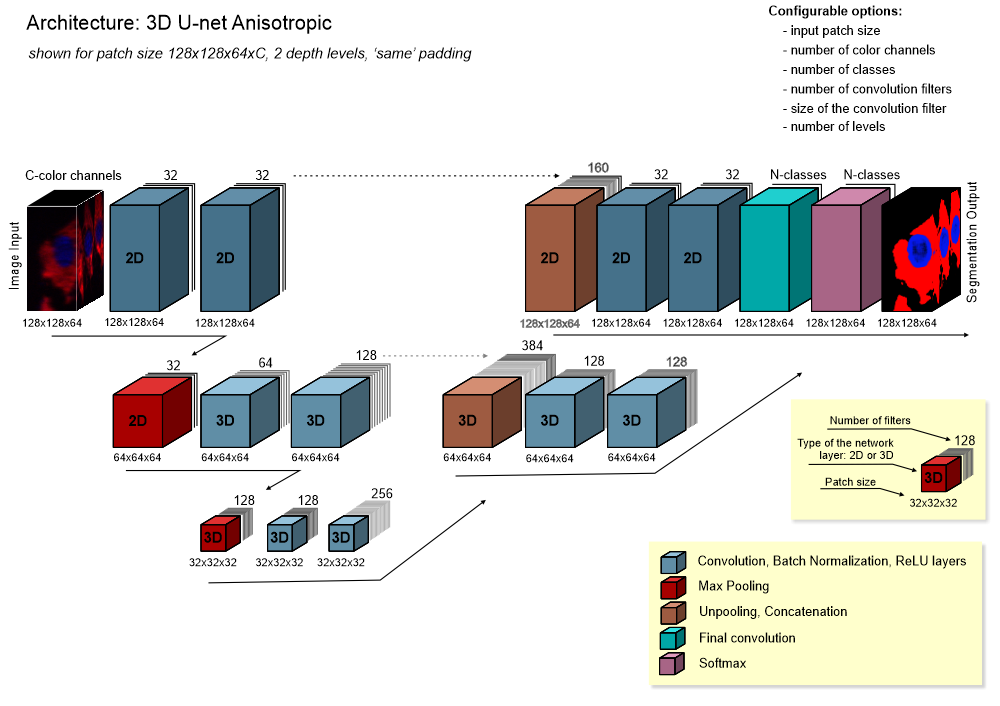
The 3D Semantic workflow suits anisotropic or slightly anisotropic 3D datasets, leveraging multiple sections to train networks for enhanced 3D structure prediction.
List of available network architectures for 3D semantic segmentation
- 3D U-net: a U-net variant for volumetric image segmentation.
References:- Cicek, Ö., et al. "3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation." MICCAI, 2016 (arXiv).
- MATLAB 3D U-Net layers.
- 3D U-net anisotropic: a hybrid of 2D and 3D U-nets, with 2D convolutions and max pooling at the top layer and 3D operations elsewhere, ideal for anisotropic voxels.
2D Patch-wise workflow
In the 2D Patch-wise workflow, training uses image patches representing specific classes. prediction processes images in blocks (with or without overlap), assigning each block a class. this is useful for quickly locating objects or targeting specific areas for semantic segmentation.
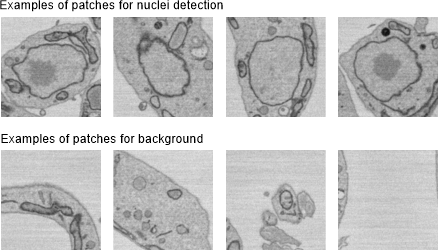
Detection of nuclei using the 2D patch-wise workflow
Examples of patches for nuclei detection:
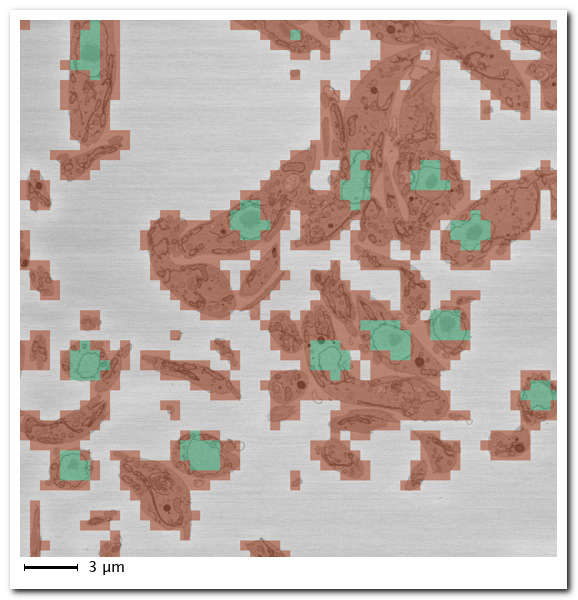
Prediction results:
- Green: predicted nuclei locations
- Red: predicted background
- Uncolored: skipped patches (dynamic masking)
Detection of spots using the 2D patch-wise workflow
Training patches for "spots" and "background":
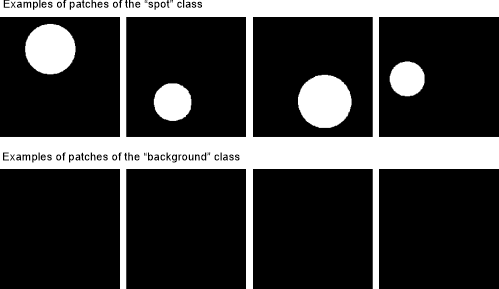
Synthetic spot detection example:
List of available networks for the 2D patch-wise workflow
Reference: - He, K., et al. "Deep Residual Learning for Image Recognition." IEEE CVPR, 2016.
References:
- He, K., et al. "Deep Residual Learning for Image Recognition." IEEE CVPR, 2016.
- Keras Resnet50.
References:
- He, K., et al. "Deep Residual Learning for Image Recognition." IEEE CVPR, 2016.
- GitHub deep-residual-networks.
Reference: - Chollet, F. "Xception: Deep Learning with Depthwise Separable Convolutions." arXiv, 2017 (arXiv).
Network filename
The button selects a file for saving or loading a network:
- In Directories and Preprocessing or Train tabs: defines the save file
- In Predict tab: loads a pretrained network for prediction
Info
The button is color-coded to match the active tab for ease of use.
GPU dropdown
The dropdown sets the execution environment:
- Name of a GPU: lists available GPUs; select one for deep learning
- Multi-GPU: uses multiple GPUs on one machine via a local parallel pool (shown if multiple GPUs are present)
- CPU only: uses a single CPU
- Parallel (under development): uses a local or remote parallel pool, prioritizing GPU workers if available
Click the button for GPU details.
Eye button
The button loads and checks the network specified in Network filename, differing from the Check network button in the Train tab, which generates a network from parameters.
Back to MIB | User interface | DeepMIB