Image Filters Panel
Back to MIB | User Guide | Panels
Overview
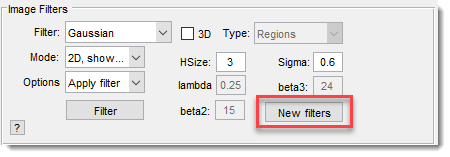
The Image Filters Panel lets you apply various 2D and 3D image filters to enhance or process your dataset.
Note on updated filters
Image filters have been significantly updated. For the best experience, use the new filters dialog via the button or Menu -> Image -> Image filters.
Image Filter
: select a filter from a list of 2D and 3D options.
Depending on the filter, specify additional parameters in edit boxes
like ,
,
,
,
,
and .
List of available filters
- Average (2D): MATLAB averaging filter. See fspecial and imfilter in MATLAB docs.
- Disk (2D): MATLAB circular averaging filter (pillbox). See fspecial and imfilter in MATLAB docs.
- DNN Denoise (2D): Denoises images using a deep neural network (MATLAB R2017b+, requires Neural Network Toolbox and a good GPU).
- Gaussian (2D): MATLAB rotationally symmetric Gaussian lowpass filter. See fspecial and imfilter in MATLAB docs.
- Gaussian (3D): Based on Dirk-Jan Kroon’s imgaussian, using multiple 1D kernels.
- Gradient (2D/3D): Generates a gradient image.
- Frangi (2D/3D): Hessian-based Frangi Vesselness filter for detecting vessels/ridges. Based on Marc Schrijver and Dirk-Jan Kroon’s implementation. References: Frangi 1998, 2001.
- Motion (2D): MATLAB filter approximating camera motion. See fspecial and imfilter in MATLAB docs.
- Median (2D): MATLAB 2D median filter for reducing "salt and pepper" noise while preserving edges. See medfilt2 in MATLAB docs.
- Median (3D): MATLAB 3D median filter (R2017a+). See medfilt3 in MATLAB docs. YouTube demo.
- Perona Malik anisotropic diffusion (2D): Smooths regions while preserving sharp gradients, by Peter Kovesi.
- Unsharp (2D): Sharpens images using unsharp masking (imsharpen, R2013a+) or contrast enhancement (fspecial/ imfilter, R2012b and older).
- Wiener (2D): MATLAB 2D adaptive noise-removal filter (wiener2), using pixel-wise Wiener methods.
- External: BMxD (2D/3D): Optional block-matching and 3D collaborative filtering. Requires separate installation (see System Requirements).
BM3D and BM4D references
- K. Dabov, A. Foi, V. Katkovnik, and K. Egiazarian, "Image denoising by sparse 3D transform-domain collaborative filtering", IEEE TIP vol. 16, no. 8, 2007;
- M. Maggioni, V. Katkovnik, K. Egiazarian, A. Foi, "A Nonlocal Transform-Domain Filter for Volumetric Data Denoising and Reconstruction", IEEE TIP, vol. 22, no. 1,pp. 119-133 2013.
Note
If is a single number, the 3D kernel
size is based on pixel size from Menu -> Dataset -> Parameters.
If two numbers (e.g., 3;3), the kernel is 3x3x3.
Mode
: choose the dataset portion to filter:
- 2D, shown slice: Filters only the current slice.
- 3D, current stack: Filters the current stack.
- 4D, complete volume: Filters the entire dataset.
Options
: select what happens post-filtration:
- Apply filter: Filters and displays the result.
- Apply and add to the image: Filters and adds the result to the original image.
- Apply and subtract from the image: Filters and subtracts the result from the original image.
Filter settings
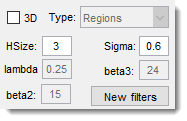
Adjust filter parameters with these edit boxes (some may be disabled based on the filter):
Filter
button: starts the filtering process.
Back to MIB | User Guide | Panels