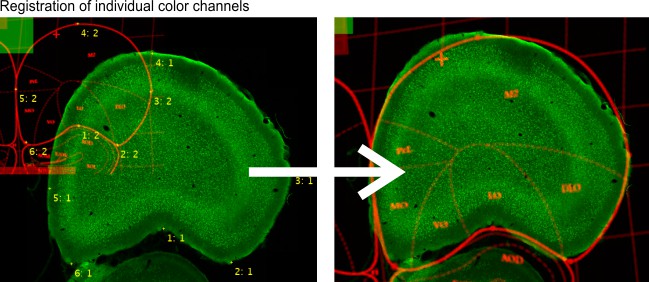
Alignment and Drift Correction
Back to MIB | User interface | Menu | Dataset
Overview
The Alignment and Drift Correction tool can be used either to align slices of the opened dataset or to align two separate datasets.
- How to do image alignment
- Multi-point landmarks
- Automatic feature-based
- Automatic feature-based using HDD mode
- HDD mode (Drift correction and Automatic feature-based only)
Current dataset panel
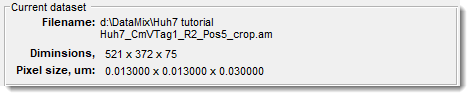
The Current dataset panel displays details of the currently opened dataset, such as its filename, dimensions, and pixel size.
Align panel
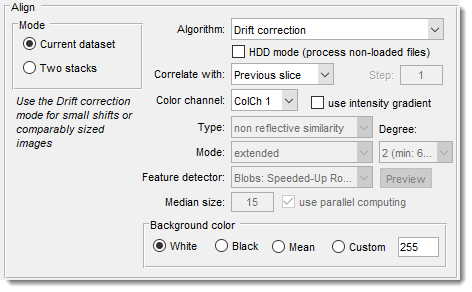
The Align panel allows selection of main parameters for alignment and drift correction.
- The Mode panel: selection of alignment mode
- Current dataset: align the opened dataset.
- Two stacks: align two stacks; the second stack can be loaded or imported from MATLAB.
- Algorithm: selection of method for alignment
Algorithm: Drift correction
Translation correction; recommended for small shifts or comparably sized images. Also implemented for image files without loading into MIB (see HDD mode).
Algorithm: Template matching
Translation correction; best for aligning two stacks when the second stack is smaller than the main stack. Not recommended for the currently opened stack.
Algorithm: Automatic feature-based version 1 and 2
Automatic alignment based on features (blobs, regions, or corners) detected on
consecutive slices. The features registered to find suitable transformation and the images are aligned using the detected transformation.
Version 2 (based on estgeotform2d)
is recommended.
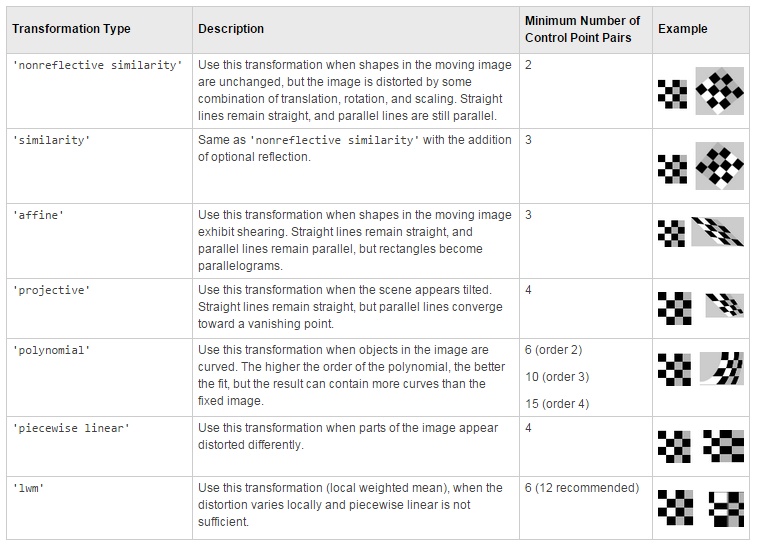
Transformations:
-
Version 1: based on estimateGeometricTransform can be used to align datasets using following transformations:
- 'similarity'
- 'affine'
- 'projective'
-
Version 2: based on estgeotform2d:
- translation: 2-D translation
- rigid: nonreflective rigid (translation + rotation)
- similarity: nonreflective similarity (translation + rotation + scaling)
- affine: general affine
(see more about various transformation types)
Results can be cropped () to the first image’s size/position or extended ().
Use to check and modify detection of points.
See matchFeatures for details.
Also implemented for HDD mode.
Feature detectors (see more on different point feature types):
- Blobs: Speeded-Up Robust Features (SURF) algorithm: robust, fast, less effective for highly detailed targets (e.g., electrical boards)
- Blobs: Detect scale invariant feature transform (SIFT): scale-invariant feature transform
- Regions: Maximally Stable Extremal Regions (MSER) algorithm: Maximally stable extremal regions (MSER) algorithm
- Corners: Harris-Stephens algorithm: Harris-Stephens algorithm. More efficient than the minimum eigenvalue algorithm
- Corners: Binary Robust Invariant Scalable Keypoints (BRISK): similar to ORB, slightly more CPU-intensive
- Corners: Features from Accelerated Segment Test (FAST): fast, extracts many keypoints, not rotation-invariant
- Corners: Minimum Eigenvalue algorithm: uses minimum eigenvalue metric to determine corner locations.
- Oriented FAST and rotated BRIEF (ORB): rotation-invariant (Oriented FAST and Rotated BRIEF), best for general use, comparable to SURF
Algorithm: AMST: Alignment to Median Smoothed Template
compensates for slight local deformations in 3D electron microscopy datasets.
Pre-align with Drift correction, then register against a median-smoothed Z copy.
Based on Hennies et al., 2020.
- : number of Z-sections for smoothing
- : configure parameters (see imregtform)
Algorithm: Single landmark point
Manual mode; mark corresponding areas on consecutive slices using the Brush tool (spot) or Annotations tool. Images are translated to align marked areas.
Algorithm: Landmarks, multi points
Align based on multiple marked points using the Selection layer or Annotations (recommended).
Corresponding points must have the same name.
Transformation types:
Algorithm: Three landmark points
Manual mode; mark three corresponding areas on consecutive slices with the Brush tool.
Images are translated/scaled/rotated to align.
Recommendation
use Landmarks, multi points instead!
Algorithm: Color channels, multi points
Register individual color channels using Annotations (Segmentation panel -> Annotations):
- annotation text identifies corresponding points
- annotation value identifies fixed (1) and transformed (2) channels
Other settings
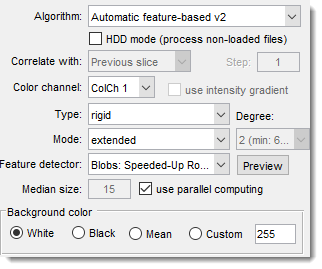
- : align files specified in
Options->HDD Mode (Image directories and formats)
without loading into MIB.
Suitable for large image collections.
Only for "Drift correction" (same size required) and "Automatic feature-based" algorithms.
: reference slice options
- Previous slice: align each slice to the previous one.
- First slice: align all to the first Z-slice; good for drift correction with minimal dataset changes.
- Relative to: align each slice to an earlier slice, defined by .
: select the color channel for alignment.
: use intensity gradients instead of raw images for better alignment.
: define background after alignment:
- White: all background pixels white.
- Black: all background pixels black.
- Mean: average value of original dataset pixels.
- Custom: specify a custom intensity.
Options panel
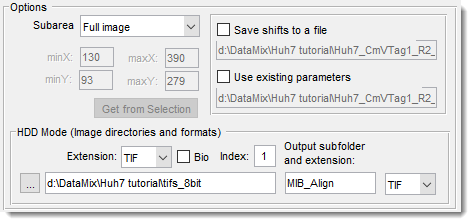
Shown only during alignment of the currently opened dataset.
: calculate correlation only from masked or selected areas.
Use the Brush tool to select distinct areas on the first and last slices,
then interpolate with I orMenu -> Selection -> Interpolate as Shape.
: speed up alignment for large uniform images. Define with , , , , or .
: save/load translation shifts to disk.
HDD Mode (image directories and formats) panel: used when HDD Mode is enabled
- : file extension to align.
- : enable Bio-Formats reader for microscope formats.
- : series index to load from a container [Bio-Formats only].
- : select directory with images.
- Output subfolder and extension: specify output subfolder (relative to input) and file format.
Second stack panel
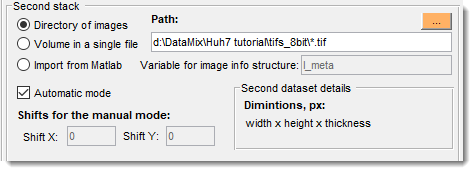
Shown only during alignment of two stacks.
- Directory of images: select directory with second stack images.
- : use if the second stack is in one file.
- : import a stack from MATLAB for alignment.
- : align automatically; otherwise, manually set and .
References and Acknowledgements
The alignment algorithm is based on:
- JC Russ, The image processing handbook, CRC Press, Boca Raton, FL, 1994.
- JD Sugar, AW Cummings, BW Jacobs, DB Robinson, A Free MATLAB Script For Spatial Drift Correction, Microscopy Today, Volume 22, Number 5, 2014. Link
Back to MIB | User interface | Menu | Dataset