Deep MIB - Predict Tab
Back to MIB | User interface | DeepMIB
Settings for efficient prediction (inference) and semantic segmentation model generation in Microscopy Image Browser.
How to start the prediction (inference) process
Prediction (inference) requires a pretrained network. if you lack one, train it first. pretrained networks can be loaded into Deep MIB for segmenting new datasets.
Steps to start prediction
- Select the pretrained network file in in the Network panel. This updates the Train panel with training settings. Alternatively, load a config file via Options tab → Config files → Load>
- Verify the prediction images directory in Directories and Preprocessing tab → Directory with images for prediction
- Confirm the output directory in Directories and Preprocessing tab → Directory with resulting images
- If needed (usually not), preprocess (convert) files:
- Set in the Directories and Preprocessing tab
- Click
- Switch to the Predict tab and press
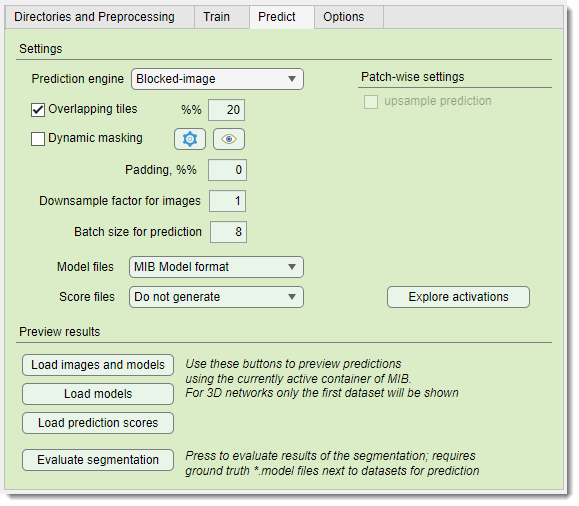
Settings section
The Settings section configures prediction parameters.
: selects the tiling engine:
- Legacy used until MIB 2.83
- Blocked-image recommended for later versions (supports Dynamic masking and 2D Patch-wise)
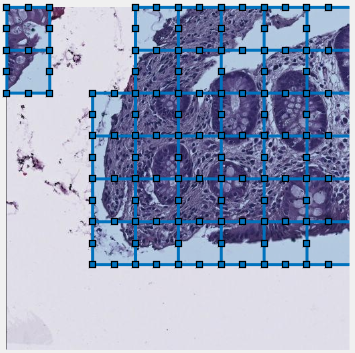
: (for Padding: same) tiles the patches with overlap to minimize edge artefacts and improve segmentation. Define overlap percentage in
Overlapping vs non-overlapping mode
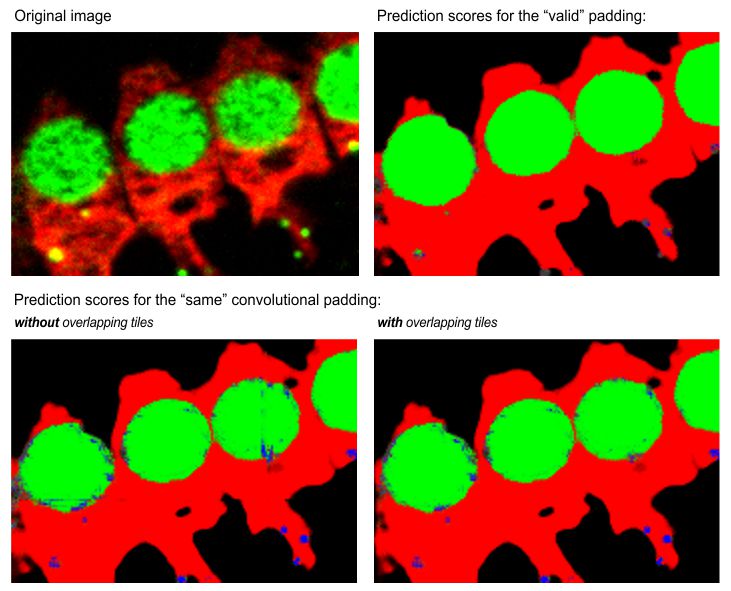
Same padding without overlap may show edge artifacts, reduced with overlapping mode.
: skips prediction on certain tiles using on-the-fly masking, configured via the  button
button
previews masking on the current Image View panel image
Dynamic masking settings and preview
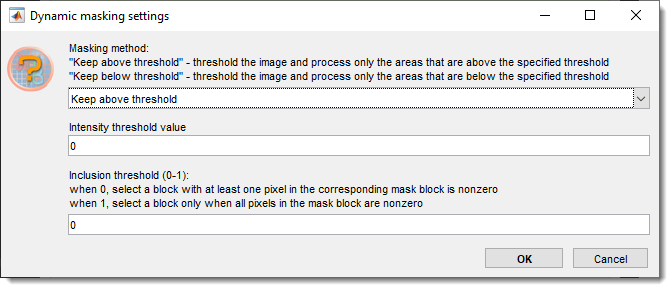
keeps blocks above or below the
sets the intensity threshold for prediction
fraction of pixels above/below threshold to keep a tile
pads images symmetrically to reduce edge artifacts
downsamples prediction images to match training
(if applicable), upsampling results to original size and in case of networks with 2 classes are also smoothed
sets the number of patches processed by GPU simultaneously, limited by GPU memory
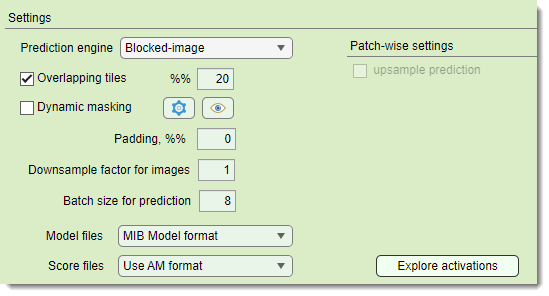
selects output format for models (CSV for patch-wise)
list of available image formats for the model files
- MIB Model format:
.modelfiles, loadable in MIB or MATLAB (model = load('filename.model', '-mat');) - TIF compressed format: LZW-compressed TIF, pixels encode classes (1, 2, 3, etc.)
- TIF uncompressed format: uncompressed TIF, pixels encode classes
selects output format for prediction score maps
list of available image formats for the score files
- Do not generate skips score files for better performance
- Use AM format AmiraMesh, compatible with MIB, Fiji, or Amira
- Use Matlab non-compressed format
.mibImg, loadable in MIB or MATLAB (model = load('filename.mibImg', '-mat');) - Use Matlab compressed format compressed
.mibImg, loadable in MIB or MATLAB - Use Matlab non-compressed format (range 0-1)
.mibImgwith 0-1 range, MATLAB-only (model = load('filename.mat');)
- : (2D patch-wise only) upsamples downsampled patch-wise predictions to match original image size
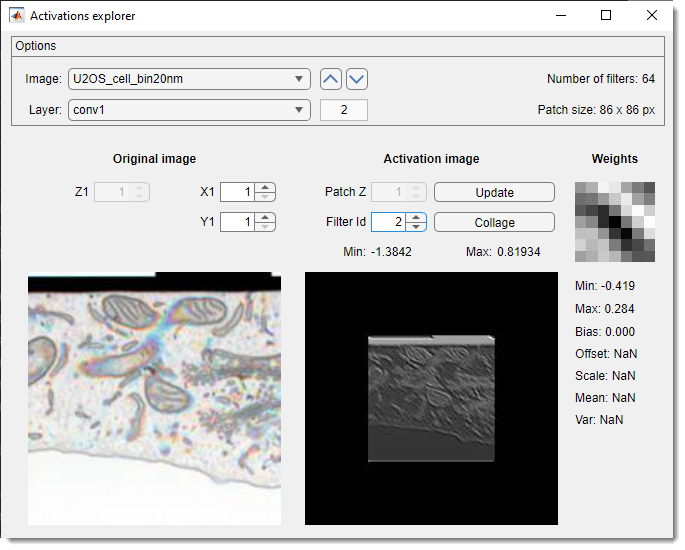
Explore activations
The Activations explorer evaluates network performance in detail. It is possible to see details of weights and activation images for all layers and filters.
- lists preprocessed prediction images. select one to load a patch matching the network’s input size. use arrows to navigate
- lists network layers. selecting a layer triggers prediction and activation image generation
- , , shifts the patch across the image. Update activations with
- adjusts Z within 3D network activation patches
- cycles through activation layers
- recalculates activations for the current patch
- creates a collage of current layer activations
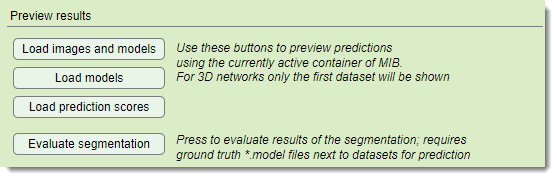
Preview results section
loads original images and segmentations
into MIB’s active buffer post-prediction
loads segmentations over the current MIB image. Requires that the image is already preloaded.
loads score images (probabilities) into the active buffer
calculates precision metrics if
ground truth labels exist in Labels under PredictionImages. Material names must match training data
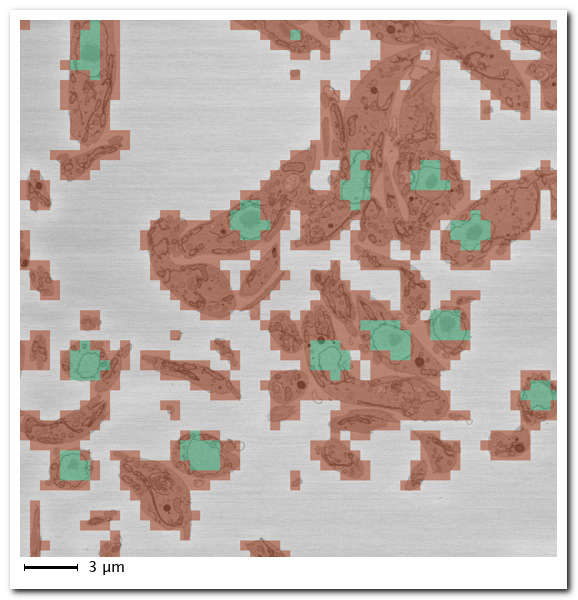
Details of the Evaluate segmentation operation
Select metrics to compute:

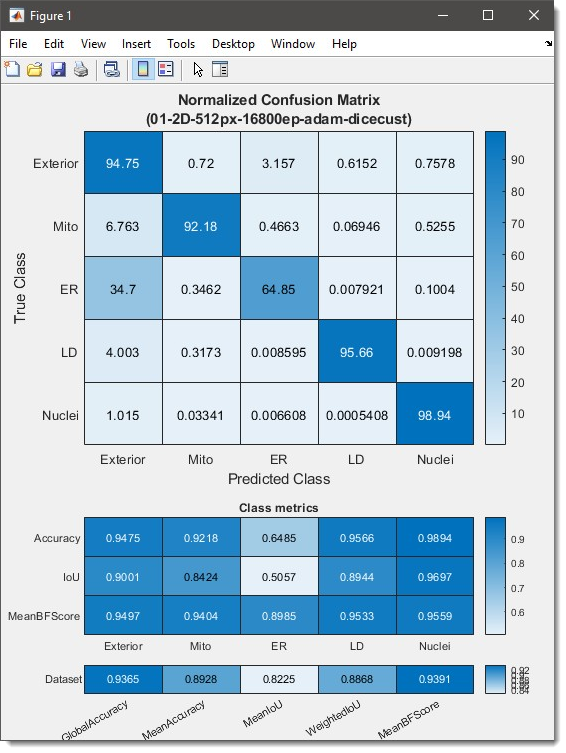
Results show a confusion matrix (0-100 scale), class metrics (Accuracy, IoU, MeanBFScore), and global metrics:
Additional metrics (label occurrence, Sørensen-Dice coefficient) are available via the dropdown:
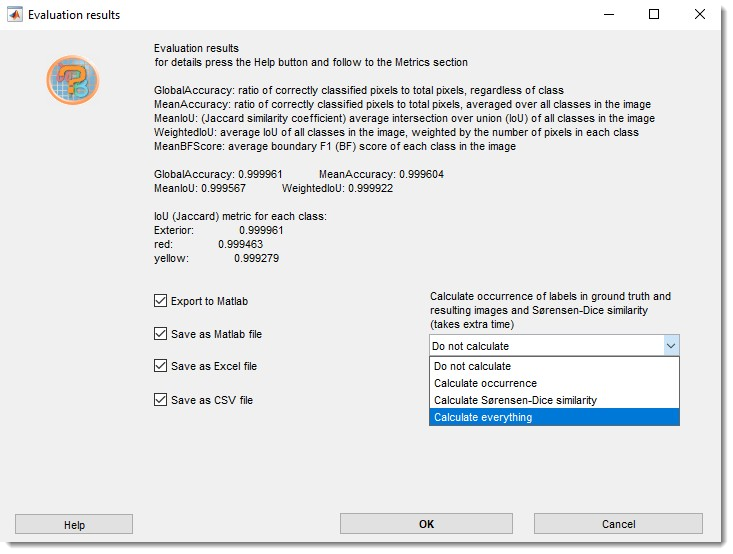
Export results to MATLAB, Excel, or CSV in 3_Results/PredictionImages/ResultsModels
(see Directories and Preprocessing). Details of the evaluation procedure are in
MATLAB evaluatesemanticsegmentation
Back to MIB | User interface | DeepMIB