Random Forest Classifier
Back to MIB | User interface | Menu | Tools
Tools for automatic image segmentation using a train-and-predict scheme based on random forest classification.
Overview
The Random Forest Classifier in Microscopy Image Browser (MIB) is a powerful method for automatic image segmentation, employing a train-and-predict approach. This implementation is based on Random Forest for Membrane Detection by Verena Kaynig and utilizes the randomforest-matlab library by Abhishek Jaiantilal. It excels in segmenting complex datasets where traditional methods like global thresholding fail due to varying background intensities.
Dataset and segmentation goal
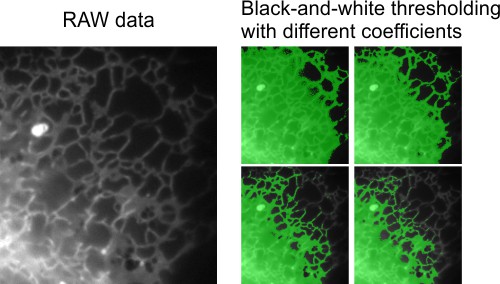
This example features a time-lapse dataset (movie) of endoplasmic reticulum captured using wide-field light microscopy. The goal is to segment the endoplasmic reticulum from the background in flat cellular regions. Global black-and-white thresholding is ineffective here due to intensity gradients across the background, making the random forest classifier a suitable alternative.
Training the classifier
Training involves manually defining regions of interest (object and background) to build a classifier that can later predict segmentation across the dataset.
- Start a new model in the Segmentation panel by clicking
- Add two materials using in the Segmentation panel
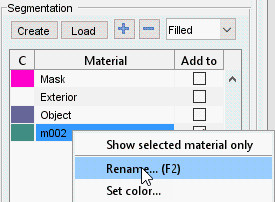
- Rename the materials:
- Highlight the first material, right-click, and select Rename from the context menu to name it Object
- Rename the second material to Background similarly
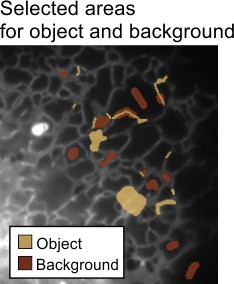
- Use the Brush tool to mark areas:
- Select endoplasmic reticulum profiles and add them to the Object material (set Add to to
1and press ) - Mark background areas and add them to the Background material (set Add to to
2and press )
- Select endoplasmic reticulum profiles and add them to the Object material (set Add to to
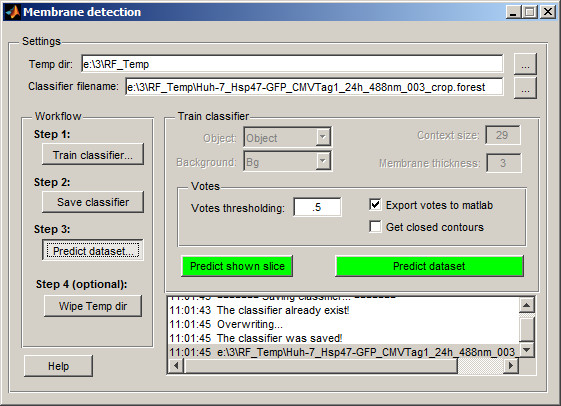
- Launch the classifier via
Menu → Tools → Classifier → Membrane detection
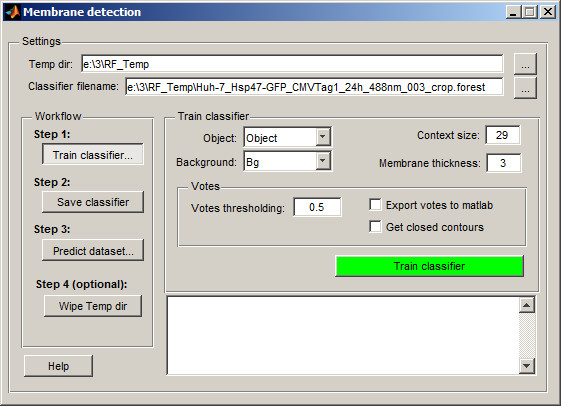
By default, the classifier creates a temporary directory (RF_Temp) next to the dataset location to store processed images and the classifier file. You can modify the temporary directory name and classifier filename in the and fields.
-
Configure the classifier:
- Set to Object
- Set to Background
- Adjust : smaller values suit short or highly curved membrane profiles
- Set : enter the approximate thickness of the membrane in pixels
- Review the Votes section: adjust the threshold, enable export to MATLAB, or enforce closed membrane profiles
-
Click to process the current slice and train the classifier based on segmented areas
- If the prediction isn’t satisfactory, segment additional areas (across multiple slices if needed) and retrain. Once satisfied, proceed to step 2 in the workflow and click
Prediction of the whole dataset
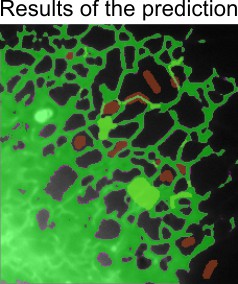
After training and saving the classifier, predict segmentation across the entire dataset.
- In the classifier window, go to step Predict dataset...
- Test the prediction on any slice by selecting it and clicking . If results are acceptable, click to process the entire dataset
The prediction results are stored in the Selection layer. Transfer them to the Model or Mask layer for further refinement or saving to disk.
Wiping the temp directory
The classifier generates large temporary files in the RF_Temp directory during prediction. Remove them by clicking or manually delete the folder using a file explorer.
Warning
Ensure you no longer need the temporary files before wiping the directory, as this action is irreversible
References
- Random Forest for Membrane Detection by Verena Kaynig
- randomforest-matlab by Abhishek Jaiantilal
Back to MIB | User interface | Menu | Tools