Directory Contents Panel
Back to MIB | User interface | Panels
Overview
This panel displays a list of files in the selected directory and provides the option to filter them based on known image and video formats.
Multiple buffer buttons
Multiple buffer buttons at the upper part of the panel provide fast access to several
datasets stored in memory. Each button represents a dataset, and the green color
indicates that a dataset is loaded in that buffer.
When you hover your cursor over a button, a tooltip with the full file name of the loaded dataset appears, helping you identify the dataset.
You can switch between datasets by clicking
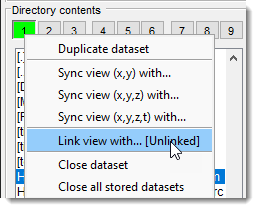
- Duplicate dataset: creates a duplicate of the currently shown dataset and places it in an available buffer.
- Sync view, (xy) with: synchronizes the view with another dataset in XY coordinates.
- Sync view, (xyz) with: synchronizes the view with another dataset in XYZ coordinates, for 4D-5D datasets.
- Sync view, (xyzt) with: synchronizes the view with another dataset in XYZT coordinates, for 5D datasets.
- Link view with: links views in two MIB buffers; shifts in one view automatically shift the other. Toggle buffers with + .
Demonstration - Close dataset: removes the selected dataset from memory.
- Close all stored datasets: removes all stored datasets from memory.
These buffer buttons and options provide convenient ways to manage and interact with multiple datasets in MIB.
File list box
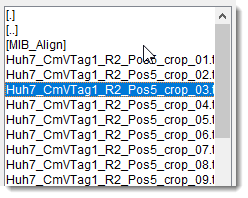
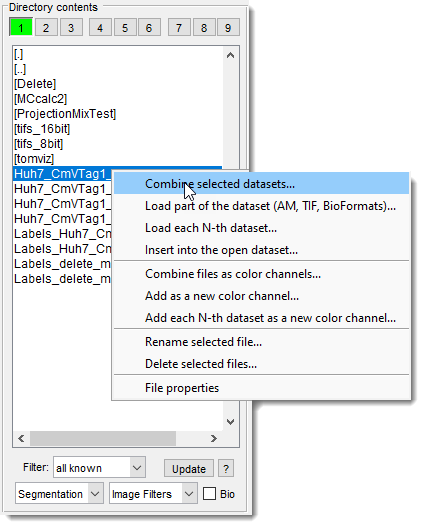
The major part of the panel is occupied by a list box that displays files in the selected directory,
chosen from the Path panel.
Files are filtered based on the specified filter.
Selecting displays all readable formats.
- Double-clicking changes the folder to the top level of the current logical drive.
- Double-clicking changes the folder to one level up.
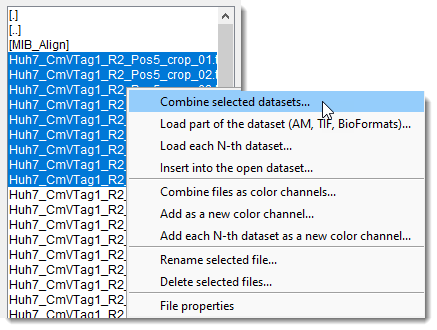
- Double-clicking navigates MIB into the clicked directory and shows files inside it
(MIB_Algnfolder in the snapshot)
- Double-
a file loads it into MIB and displays it in the Image View panel. - Select individual datasets by holding or
and clicking
on files. Load them via the context menu ( options below).
The context menu, accessed with
- Combine selected datasets: combine selected datasets into a single 3D stack.
- Load part of the dataset (AM, TIF, BioFormats): load a specific part of a larger dataset, defining start/end points, z-step, and XY binning (available for Amira Mesh, TIF, and BioFormats-readable datasets) demo.
- Load each N-th dataset: assemble every N-th file into a 3D stack.
- Insert into the open dataset: insert selected files into the currently open dataset.
- Combine as color channels: combine selected datasets into a single 2D slice, each assigned to a color channel.
- Add as a new color channel: add images as a new color channel to the existing dataset. Select channels in the View Settings panel.
- Add each N-th dataset as a new color channel: add images with an N-step as a new color channel to the existing dataset.
- Rename selected file: rename the selected file.
- Delete selected files: permanently delete selected files from the disk.
- File properties: show file details like date/time and size in bytes.
Note
Filter
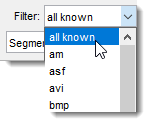
The dropdown allows filtering the file list
based on available image type filters.
Available extensions depend on whether the standard
or Bio-Formats reader is used and if
virtual mode is enabled, resulting in four filter configurations.
Modify extensions in the filter list
To modify file extensions the list of filters:
click over . - Choose an option:
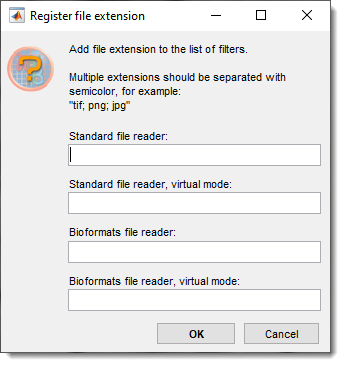
- Register extension: add extensions to the filter list.
- Remove selected extension: remove a selected extension from the filter list.
Update button
The button refreshes the file list in the Directory Contents panel.
Left interchangeable panels
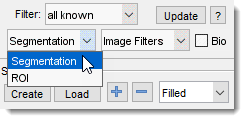
The selected panel appears in the lower-left part of MIB:
Right interchangeable panels
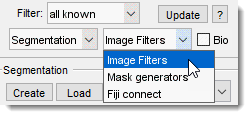
The selected panel appears in the bottom-right part of MIB:
Bio checkbox
When is checked,
the Bio-Formats reader loads datasets.
MIB supports specific biological image formats (native to various microscopes) via the
Bio-Formats Java library, stored in
the MIB\ImportExportTools\BioFormats directory.
Note
Performance of the Bio-Formats reader may be slower than standard native readers due to Java-related overheads.
Help button
The button links to this help page.
Back to MIB | User interface | Panels