Model Menu
Back to MIB | User interface | Menu
Overview
Actions that can be applied to the Model layers. The Model layer is one of three main segmentation layers (Model, Selection, Mask) which can be used in combination with other layers. See more about segmentation layers in the Data layers section.
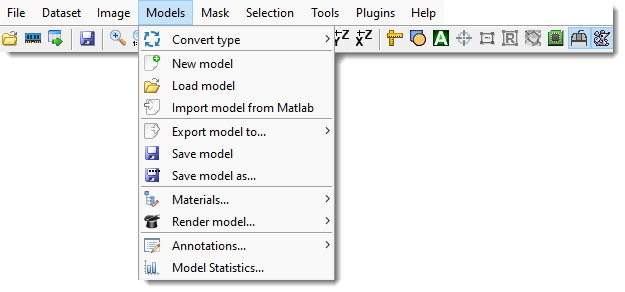
Convert type
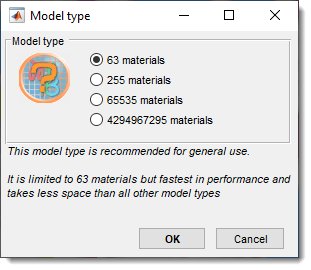
Convert the model to a different type; the current type is checked in the menu.
- 63 materials (default): Stores Models, Selection, and Mask layers in a single memory container, reducing memory requirements and improving performance, but limits materials to 63.
- 255 materials: Allows up to 255 materials, requiring additional memory for Selection and Mask layers (doubles memory usage).
- 65535 materials: Allows up to 65535 materials, requiring ~1.25x more memory than 255 materials. The Segmentation panel appearance changes in this mode.
Short demonstration - 4294967295 materials: Allows up to 4294967295 materials, requiring twice the memory of 65535 materials.
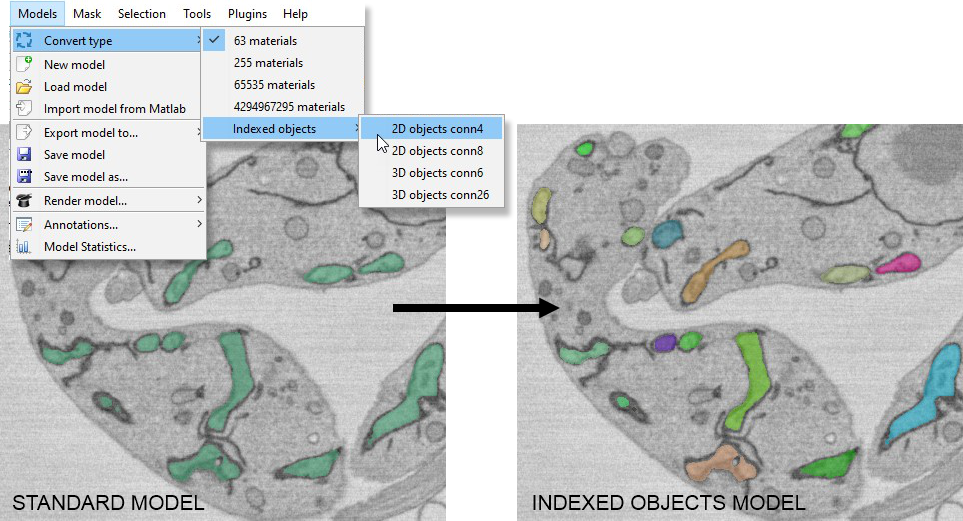
- Indexed objects → 2D objects conn4: Detects all 2D objects (connectivity 4) in all materials and generates a new model where each object has a unique index.
- Indexed objects → 2D objects conn8: Detects all 2D objects (connectivity 8) in all materials and generates a new model with unique indices.
- Indexed objects → 3D objects conn6: Detects all 3D objects (connectivity 6) in all materials and generates a new model with unique indices.
- Indexed objects → 3D objects conn26: Detects all 3D objects (connectivity 26) in all materials and generates a new model with unique indices.
How to work with models having more than 255 materials
 Materials should be named with numbers representing the current working material index
(e.g., 11555 means that when selection is added to the model, it will be assigned to index 11555).
Materials should be named with numbers representing the current working material index
(e.g., 11555 means that when selection is added to the model, it will be assigned to index 11555).
Select materials by:
- Right-clicking the segmentation table and choosing Rename... or by pressing F2
- Hovering over an object in the Image View panel and pressing Ctrl+F.
New model
Allocates space for a new model. Use this to start a new model or delete the existing one.
Alternatively, use the button in the Segmentation Panel.
For model types see above
Load model
Loads a model from disk. By default, MIB reads models in MATLAB format (.model), but other formats are supported.
- .AM, Amira Mesh: Amira Mesh label field for models from Amira.
- .NRRD, Nearly Raw Raster Data: Compatible with 3D Slicer.
- .MRC, Medical Research Council format: Compatible with IMOD. Can load multiple MRC files, each encoding an object, and merge them into a single model.
- .PNG, PNG format: Saves models as 2D slices in Portable Network Graphic format.
- .TIF, TIF format: Saves models as 2D slices or 3D volumes in Tag Image File format.
Note
Almost any standard image format can be loaded as a model using the All files (.) filter in the Open model dialog.
Alternatively, use the button in the Segmentation Panel.
Note
Models can be opened by drag-and-droping of the selected model files into the Segmentation panel
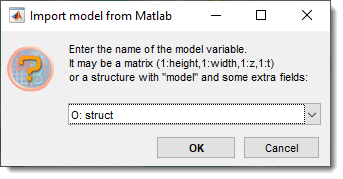
Import model from MATLAB
Imports a model from the main MATLAB workspace.
Provide a variable name with a matrix matching the dataset dimensions [1:height, 1:width, 1:no-slices] of uint8 class, or a structure with the fields below.
- .model: Matrix matching dataset dimensions
[1:height, 1:width, 1:depth, 1:time]ofuint8class. - .modelMaterialNames (optional): Cell array with material names.
- .modelMaterialColors (optional): Matrix with colors (0-1)
[1:materialIndex, Red Green Blue]. - .labelText (optional): Cell array with annotation labels.
- .labelPosition (optional): Matrix with annotation positions
[1:annotationIndex, x y z].
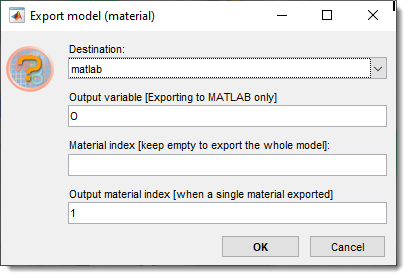
Export model to...
Exports the model from MIB to other programs.
- MATLAB: Exports to the main MATLAB workspace as a structure (see above). Can be re-imported using Import model from MATLAB.
- Imaris as volume: Exports to Imaris if available. See System Requirements for details.
Save model
Saves the model to a file in MATLAB format without prompting for a filename.
- Default template:
Labels_NAME_OF_THE_DATASET.model. - Otherwise, the filename is taken from last Save model as... opetation
- Otherwise, the filename is assinged when a model is loaded .
Info
Can also be saved using the Save model button in the Toolbar.
Save model as...
Prompts for a filename and format to save the model.
- .AM, Amira Mesh: RAW, RAW-ASCII, or RLE compressed formats (RLE is slow).
- .MAT, MATLAB format: Native format for MIB version 1.
- .MODEL, MATLAB format (default): Native format for MIB version 2.
- .MOD, IMOD format: Contours for IMOD.
- .MRC, IMOD format: Volume for IMOD.
- .NRRD, Nearly Raw Raster Data: Compatible with 3D Slicer.
- .PNG: 2D slices in Portable Network Graphic format.
- .STL, STL format: Triangulated mesh for visualization programs like Blender.
- .TIF, TIF format: 2D slices or 3D volumes.
Material...
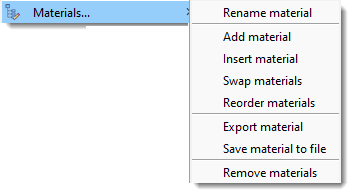
Operations for model materials, also available by right-clicking the Segmentation table.
- Rename material: Rename the selected material.
- Add material: Add a new material to the bottom of the list.
- Insert material: Insert a material at a specified position, shifting others.
- Swap materials: Swap positions of two materials.
- Reorder materials: Reorder materials with a new sequence.
- Export material: Export the selected material to MATLAB or Imaris.
- Save material to file: Save the selected material to a file.
- Remove material: Remove selected material(s) from the model.
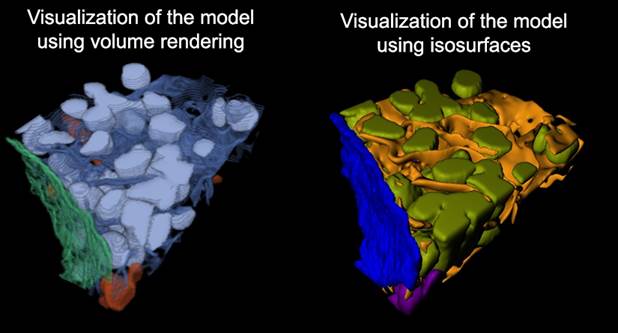
Render model...
Renders segmented models using various methods.
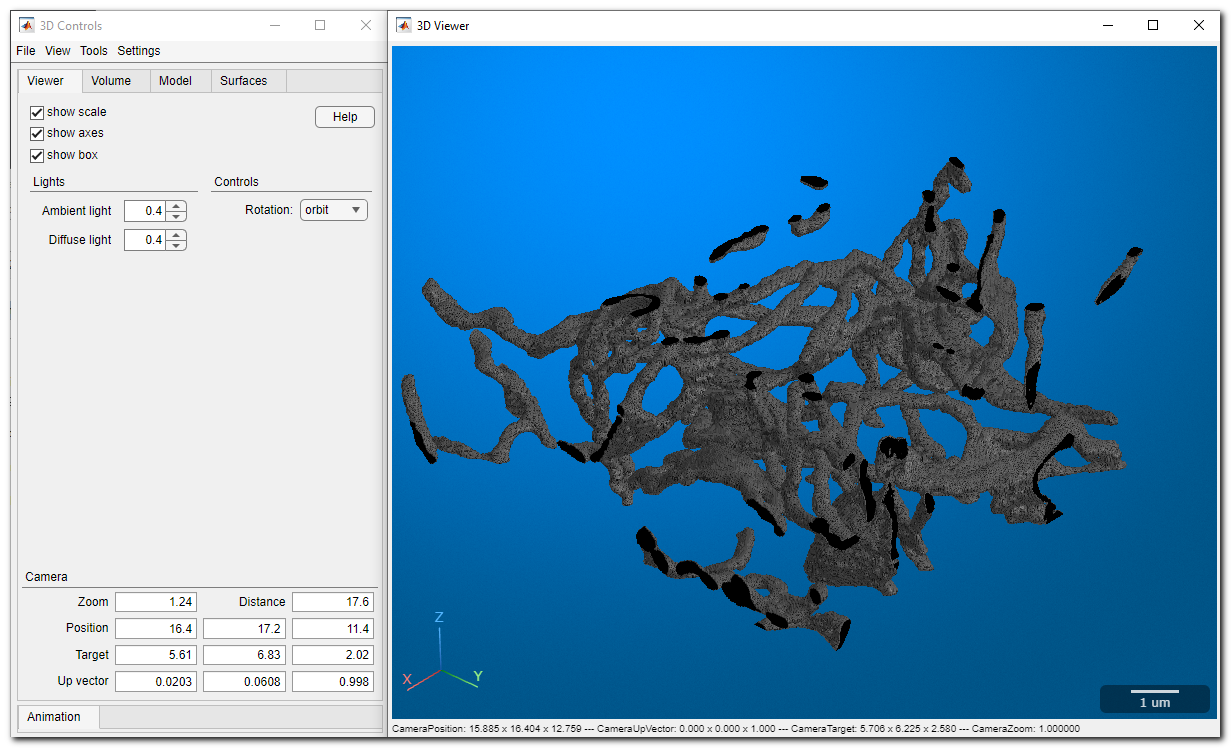
MIB rendering
From MIB 2.5 and MATLAB R2018b, materials can be visualized in MIB with hardware-accelerated volume rendering. Datasets can be downsampled. Snapshots and animations are supported.
See more: MIB 3D Viewer
Introduction to an updated 3D viewer
Original version of 3D viewer
MATLAB isosurface
Uses MATLAB to generate and visualize isosurfaces with a modified view3d function by Torsten Vogel.
- Double
to restore the original view. - Z: Switch from ROTATION to ZOOM.
- to ZOOM. - hold middle mouse to PAN.
- R: Switch from ZOOM to ROTATION.
- ROTATION:
for xy-axis rotation, middle mouse for z-axis rotation.
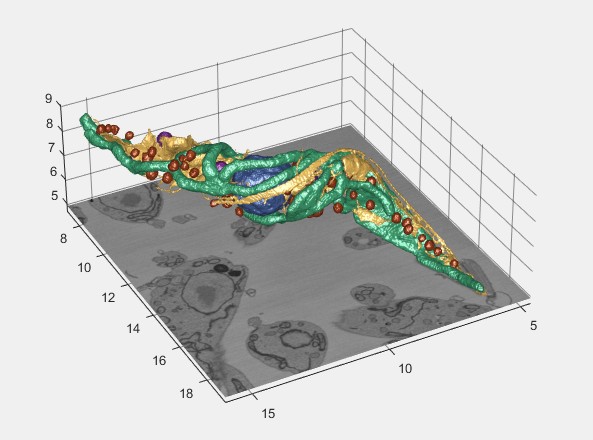
MATLAB isosurface and export to Imaris
Generates isosurfaces in MATLAB and exports them to Imaris for visualization.
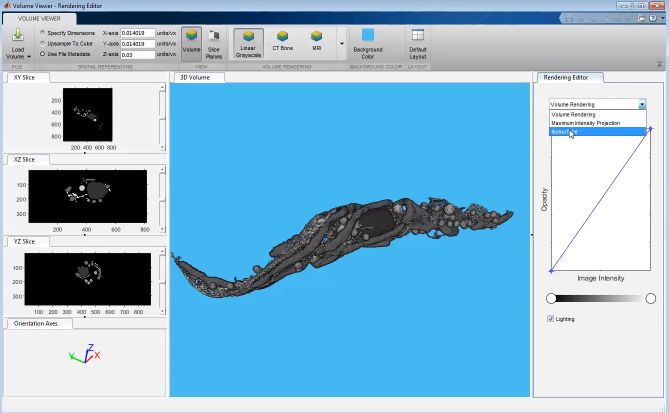
MATLAB volume viewer
Renders the model using MATLAB’s Volume Viewer (R2017b–R2019b+). In R2019b, materials can display with the volume, but lighting controls are limited.
Fiji volume
Uses Fiji 3D Viewer for volume visualization.
Requires Fiji installation (see System Requirements).
Imaris surface
Renders the model in Imaris. Requires Imaris and ImarisXT (see System Requirements).
The rendered material is specified in the Material list of the Segmentation Panel.
Annotations...
Modify the Annotations layer.
- List of annotations...: opens a window with existing annotations (see Segmentation Tools).
- Export to Imaris as Spots: exports annotations as Spots in Imaris (export the dataset first).
- Remove all annotations...: deletes all annotations in the model.
Model Statistics...
Gets statistics for the selected material, usable to filter the model by object properties.
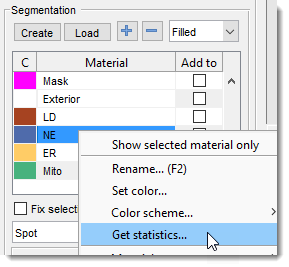
Accessible also via
Segmentation Panel → Materials List → Right-click → Get statistics...
See Mask and Model Statistics for details.
Back to MIB | User interface | Menu