File Menu
The File menu in Microscopy Image Browser (MIB) provides tools for handling image datasets, including importing, exporting, saving, and rendering options. This page details each submenu entry, with demonstrations and references where available.
Back to MIB | User interface | Menu
File Menu Overview
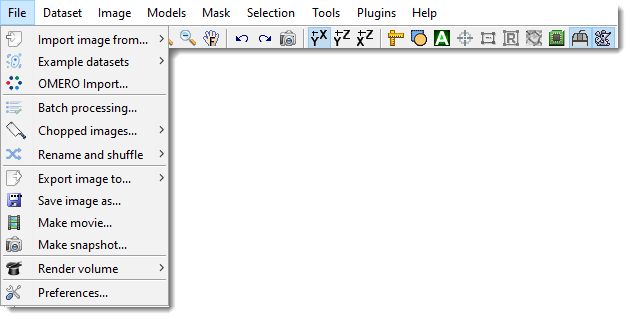
The image above shows the File menu dropdown, located in the menu bar at the top of the MIB interface. This menu organizes essential file-related actions for working with multidimensional microscopy datasets.
Import Image From
Load images into MIB from various sources:
- MATLAB: Import an image from the main MATLAB workspace into MIB
Optionally include acontainers.Mapwith dataset parameters that were generated by the Export image to command Brief demo - Clipboard: Paste an image from the system clipboard
Uses the IMCLIPBOARD function by Jiro Doke, MathWorks, 2010 Brief demo - Imaris: Import a dataset from Imaris
Requires Imaris and ImarisXT and uses IceImarisConnector by Aaron C. Ponti, ETH Zurich
Demo - URL: Open an image from a URL address
The link must include the protocol (e.g.,http://) Brief demo
Example Datasets
Quickly access demo datasets and full DeepMIB projects for image segmentation, grouped by data collection techniques.
DeepMIB projects → Synthetic 2D Large Spots
A complete DeepMIB project with a synthetic dataset for testing semantic segmentation
Includes a trained DeepLabV3-Resnet18 network for detecting large spots on a black background
Load the network via 2D_LargeSpots_2cl_DeepLabV3.mibCfg using:
Menu → Tools → Deep learning segmentation → Options tab → Config files → Load
| Directory Tree | Segmentation Example |
|---|---|
 |
 |
Reference: DOI: 10.5281/zenodo.10203188
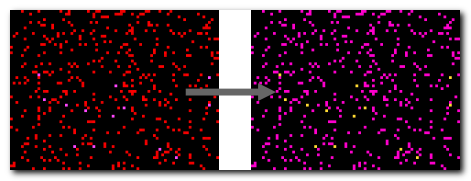
DeepMIB projects → Synthetic 2D Small Spots
A complete DeepMIB project with a synthetic dataset for testing semantic segmentation
Includes a trained U-net network for detecting small random spots of two colors on a black background
Load the network via 2D_SmallSpots_3cl_Unet.mibCfg using:
Menu → Tools → Deep learning segmentation → Options tab → Config files → Load
| Directory Tree | Segmentation Example |
|---|---|
 |
 |
Reference: DOI: 10.5281/zenodo.10203764
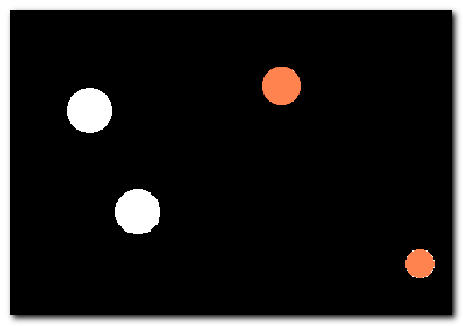
DeepMIB projects → Synthetic 2.5D Large Spots
A complete DeepMIB project with a synthetic dataset for testing 2.5D depth-to-color semantic segmentation
Includes trained 2.5D DeepLabV3-Resnet18 and U-net networks for segmenting large 3D spots (ignoring 2D spots) using 5-slice subvolumes
Load the networks via:
- Spots_25D_DLv3RN18_Z2C_xy200z5 (DeepLabV3-based)
- Spots_25D_Unet_Z2C_xy200z5 (U-net-based)
Using Menu → Tools → Deep learning segmentation → Options tab → Config files → Load or drag-and-drop the config file into DeepMIB
| Directory Tree | Segmentation Example |
|---|---|
 |
 |
Reference: DOI: 10.5281/zenodo.10212417
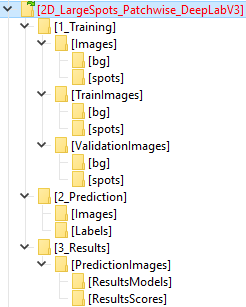
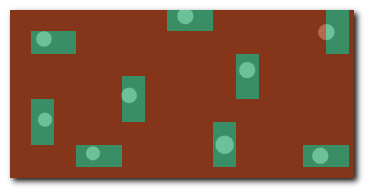
DeepMIB projects → Synthetic 2D Patch-wise
A complete DeepMIB project with a synthetic dataset for testing patch-wise segmentation
Includes a trained Resnet18 network for detecting patches of large white spots on a black background
Load the network via 2D_LargeSpots_Patchwise_Resnet18.mibCfg using:
Menu → Tools → Deep learning segmentation → Options tab → Config files → Load
| Directory Tree | Segmentation Example |
|---|---|
 |
 |
Reference: DOI: 10.5281/zenodo.10203861
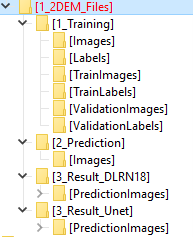
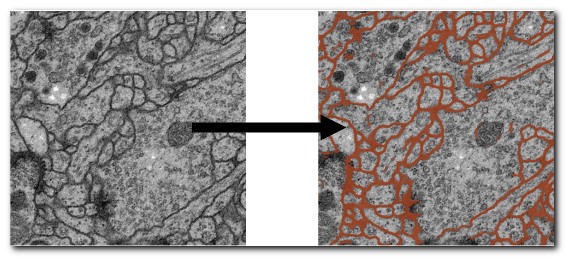
DeepMIB projects → 2D EM Membranes
A complete DeepMIB project for segmenting membranes from serial-section TEM images
Includes trained U-net and DeepLabV3-Resnet18 networks
Updated project from DeepMIB paper (Figure 1a)
Load via 2D_EM_membranes_Unet.mibCfg or 2D_EM_membranes_DLabRN18.mibCfg using:
Menu → Tools → Deep learning segmentation → Options tab → Config files → Load
See readme.txt for details
| Directory Tree | Segmentation Example |
|---|---|
 |
 |
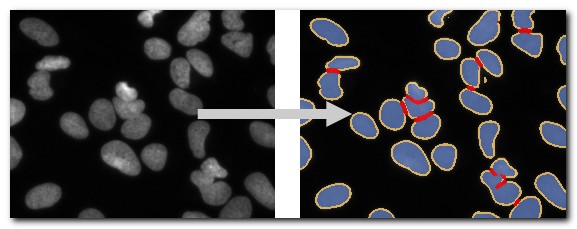
DeepMIB projects → 2D LM Nuclei
A complete DeepMIB project for segmenting nuclei, boundaries, and touching edges
Includes a trained U-net network
Updated project from DeepMIB paper (Figure 1b)
Load via valid_252px_32patches_50ep.mibCfg using:
Menu → Tools → Deep learning segmentation → Options tab → Config files → Load
See readme.txt for details
| Directory Tree | Segmentation Example |
|---|---|
 |
 |
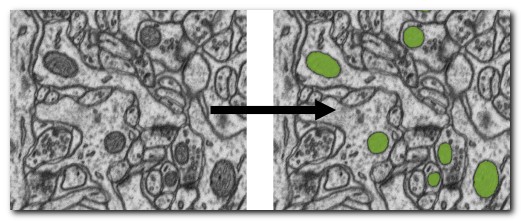
DeepMIB projects → 3D EM Mitochondria
A complete DeepMIB project for segmenting mitochondria
Includes a trained 3D U-net network
Updated project from DeepMIB paper (Figure 1c)
Load via NoValidation_valid_Aug_128px_256pat.mibCfg using:
Menu → Tools → Deep learning segmentation → Options tab → Config files → Load
See readme.txt for details
| Directory Tree | Segmentation Example |
|---|---|
 |
 |
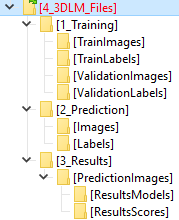
DeepMIB projects → 3D LM Inner Hair Cells
A complete DeepMIB project for segmenting inner hair cells, nuclei, and synapses
Includes a trained 3D anisotropic U-net network
Updated project from DeepMIB paper (Figure 1d)
Load via InnerEar3D_Hybrid_Same_136x64px_120ep.mibCfg using:
Menu → Tools → Deep learning segmentation → Options tab → Config files → Load
See readme.txt for details
| Directory Tree | Segmentation Example |
|---|---|
 |
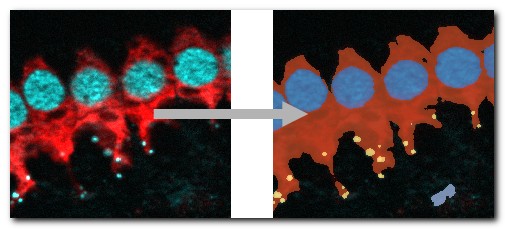 |
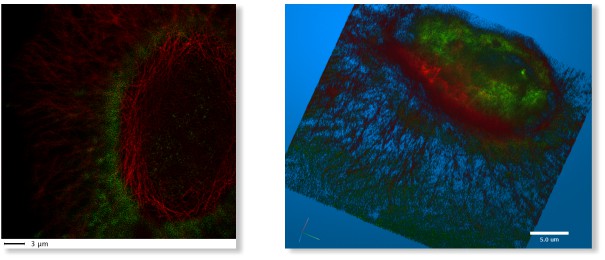
LM → 3D SIM ER
A 3D super-resolution structured illumination light microscopy dataset of endoplasmic reticulum
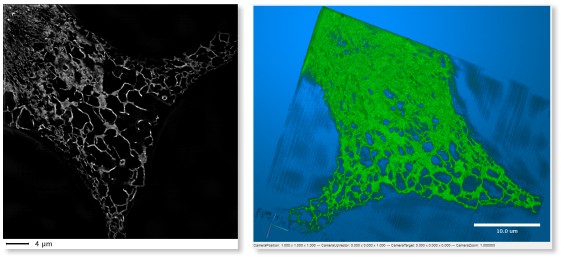
LM → WF ER Photobleaching
A wide-field time-lapse imaging dataset of endoplasmic reticulum with visible photobleaching effect
SBEM → Huh-7 and Model
A small fragment of serial block face scanning electron microscopy dataset featuring a Huh-7 cell and a model of nuclei, endoplasmic reticulum, mitochondria, and lipid droplets

SBEM → Trypanosoma and Model
A fragment of serial block face scanning electron microscopy dataset featuring a Trypanosoma brucei cell and a model of nuclei, endoplasmic reticulum, mitochondria, vesicles, lipid droplets, and cytoplasm

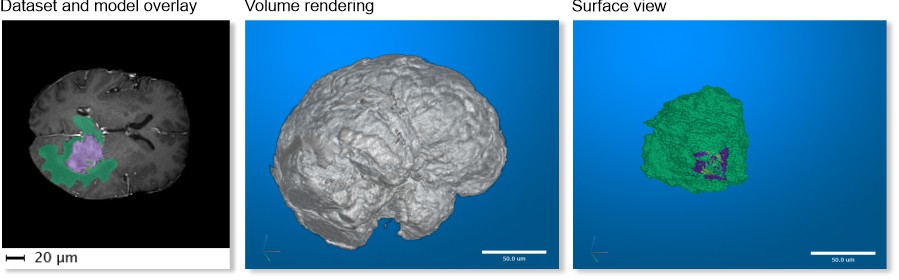
MRI → MATLAB Brain and Model
A test brain dataset from MATLAB, captured with magnetic resonance imaging (MRI), including a tumor model
Available only in MIB for MATLAB
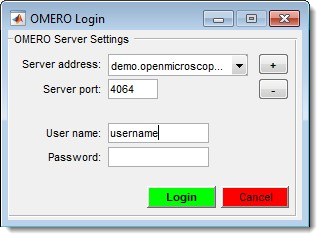
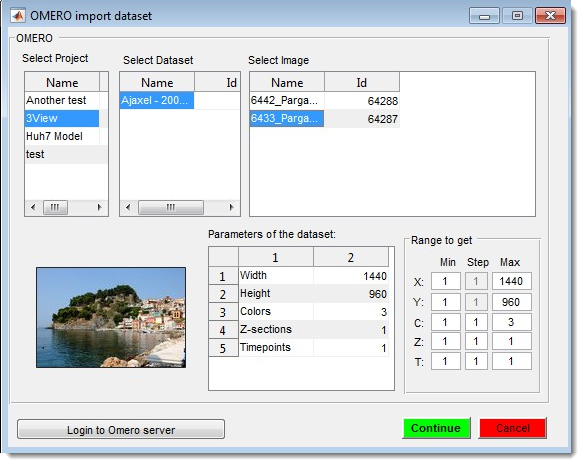
OMERO Import
Connect to an OMERO server and load images. Requires OMERO server files; see System Requirements for installation details.
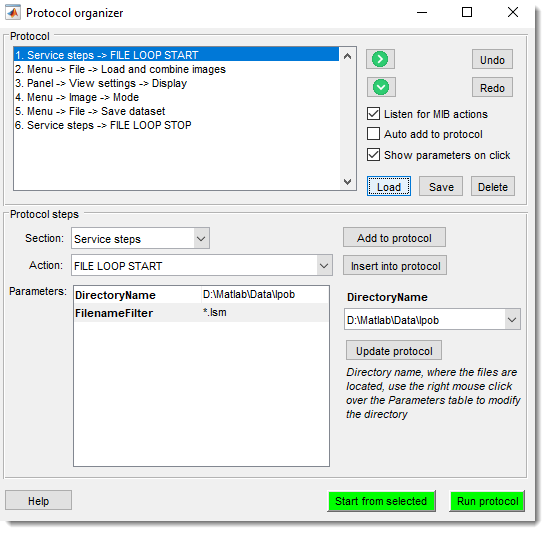
Batch Processing
Batch processing dialog.
Design and apply image processing workflows to multiple images automatically.
See Batch Processing for details.
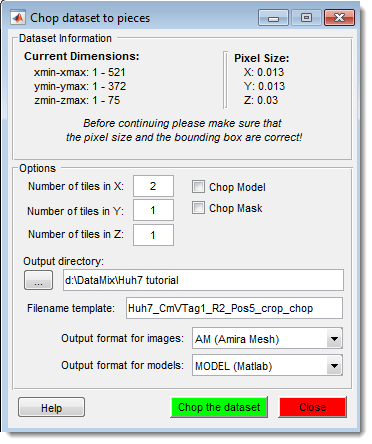
Chopped Images
Split a large dataset into smaller parts and recombine them later, or fuse previously cropped datasets.
See Chopped Images for details.
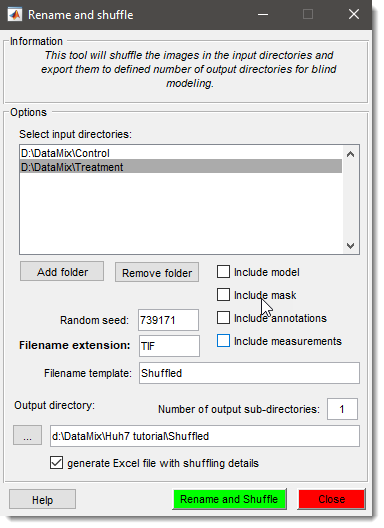
Rename and Shuffle
Shuffle files for blind modeling and revert models to original filenames for analysis.
See Rename and Shuffle for details.
Export Image To
Export images to external applications:
- MATLAB: Export includes a
containers.Mapwith dataset parameters for re-import into MIB Demo - Imaris: Export to Imaris (requires Imaris installation)
Save Image As
Save the open dataset to disk in various formats:
Supported Image Formats
- AM, Amira Mesh: Binary format for Amira Mesh
- JPEG, Joint Photographic Experts Group: Lossy compressed RGB format
- HDF5, Hierarchical Data Format: Version 5 format
- MRC, MRC format for IMOD: Compatible with IMOD
- NRRD, Nearly Raw Raster Data: Compatible with 3D Slicer
- OME-TIFF 5D (*.ome.tiff): 5D stack using BioFormats library
- PNG, Portable Network Graphics (*.png): Lossless format
- TIF format, LZW compressed: Multilayered or sequence of 2D files (max 2GB due to 32-bit offsets)
- TIF format, non-compressed: Same as above, uncompressed
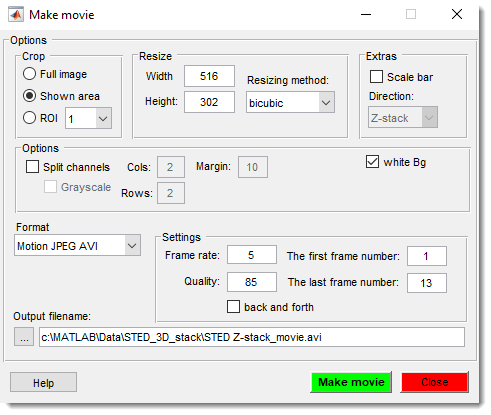
Make Movie
Save the dataset as a movie file, capturing all visible objects in the image view window.
See Make Movie for details.
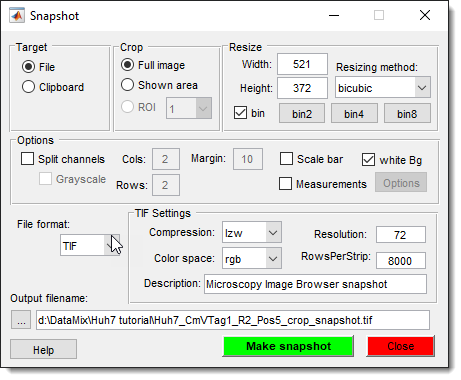
Make Snapshot
Capture a snapshot of the current slice, including all visible objects.
See Make Snapshot for details.
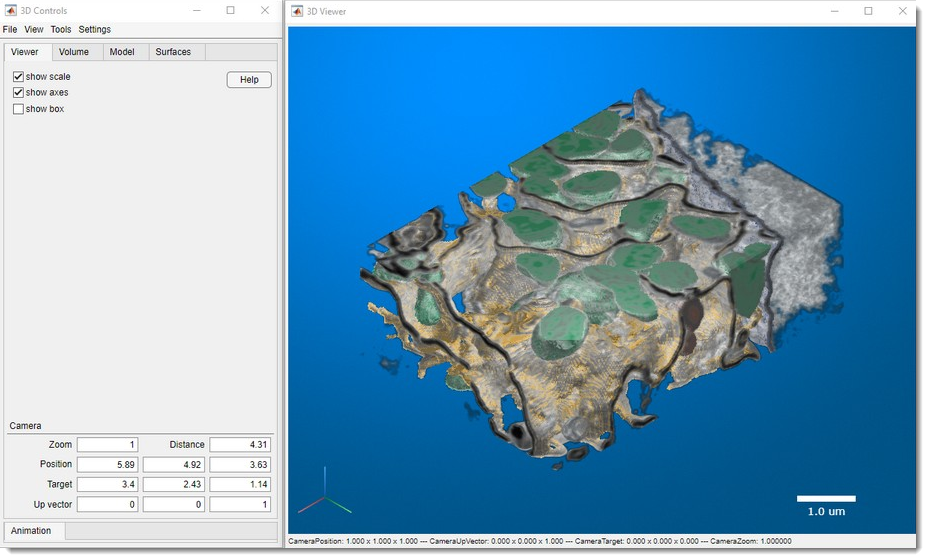
Render Volume
Visualize volumes in 3D using three methods:
MIB Rendering (recommended)
Hardware-accelerated volume rendering in MIB (since version 2.5, MATLAB R2018b).
Supports downsampling, snapshots, and animations. Updated in MIB 2.84 (MATLAB R2022b) to render 1-3 color channels with models (MATLAB-only as of 2.84).
See 3D Viewer for details.
Volume Rendering Engines of MIB
Select the engine in:
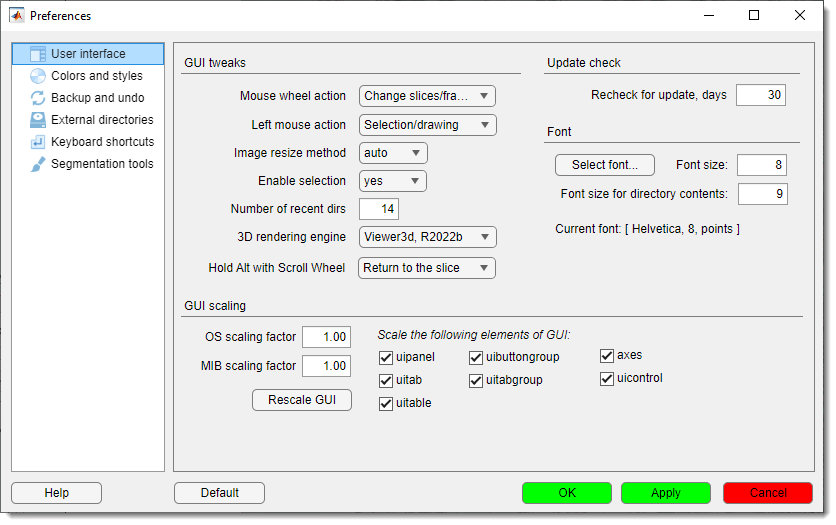
MIB → Menu → File → Preferences → User interface → 3D rendering engine
Demo
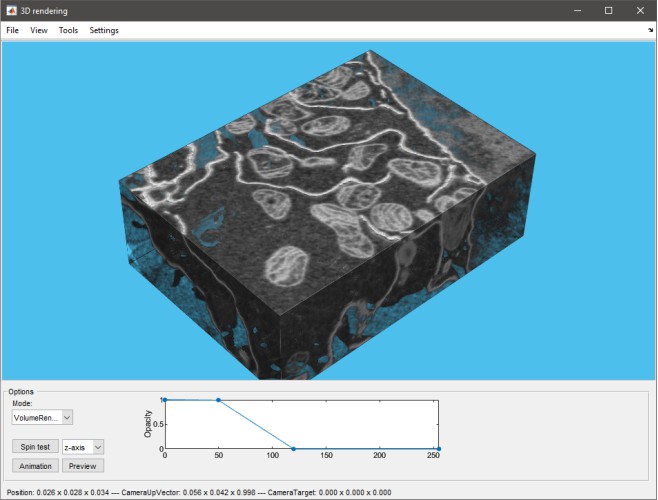
MATLAB Volume Viewer
Export to MATLAB’s Volume Viewer app (R2017a or newer, not available in compiled MIB).
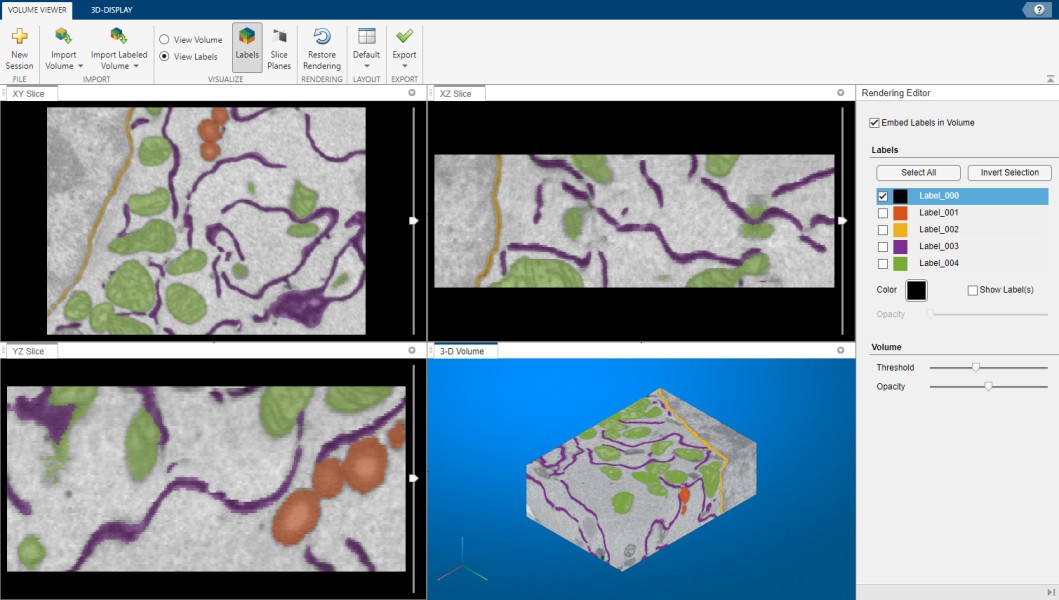
Demo
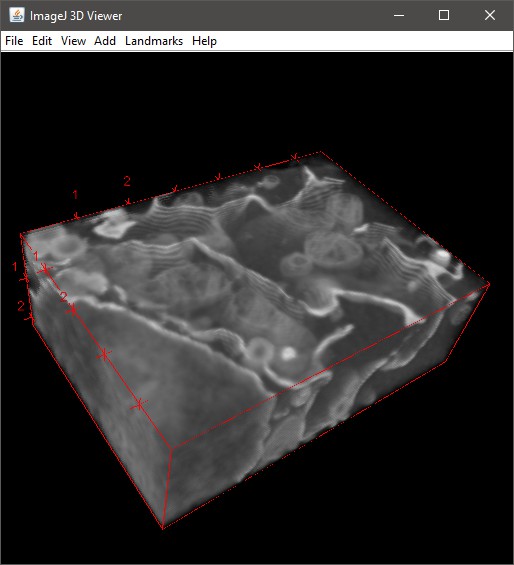
Fiji 3D Viewer
Render via Fiji (requires installation; see System Requirements). Demo
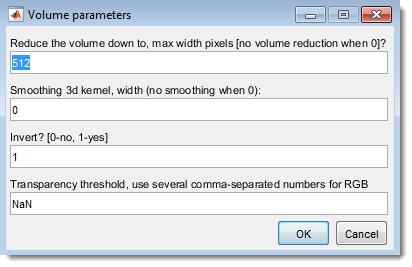
- Reduce the volume down to, max width pixels: Resize dataset (0 for no resizing)
- Smoothing 3D kernel, width: Apply Gaussian blur (0 for no smoothing)
- Invert? [0-no, 1-yes]: Invert for electron microscopy
- Transparency threshold: Set transparency (comma-separated for each channel), adjustable in Fiji’s 3D Viewer (Edit → Attributes → Adjust threshold)
Preferences
Edit MIB preferences, including colors for Selection, Model, and Mask layers, mouse wheel behavior, key settings, and Undo options.
See Preferences for details.
MIB saves configuration in a file generated upon closing:
Configuration File Location
- Windows:
C:\Users\Username\MATLAB\mib.mator TEMP directory (C:\Users\Username\AppData\Local\Temp\)
Access TEMP via Windows → Start → %TEMP% - Linux:
/home/username/Matlabor local TEMP directory - MacOS:
/Users/username/Matlabor local TEMP directory
Location of the configuration file is shown upon MIB startup as:
MIB parameters file: C:\Users\username\Matlab\mib.mat
Back to MIB | User interface | Menu