Watershed Segmentation
Back to MIB | User interface | Menu | Tools
Semi-automated image segmentation using the Watershed method in Microscopy Image Browser.
Overview
The Watershed segmentation tool in MIB provides semi-automated segmentation using the Watershed method, ideal for separating objects with distinct boundaries, such as membrane-enclosed organelles.
Note
For better interactivity and performance consider using the Graphcut segmentation tool instead.
Mode panel
The Mode panel lets you select the segmentation scope.
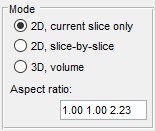
- segments only the current slice in the Image View panel
- applies 2D segmentation to each slice individually
- performs 3D segmentation on the entire dataset or a subarea (see Subarea panel below)
- displays the dataset’s aspect ratio, derived from voxel sizes in (Menu → Dataset → Parameters). this is used when Watershed relies on a distance map (see Image segmentation settings below)
Subarea panel
The Subarea panel defines a dataset subset for processing, useful for large datasets or binning.
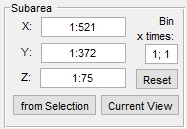
- sets the width range (e.g.,
1:512) - sets the height range
- sets the z-slice range
- fills X, Y, and Z with coordinates from the Selection layer’s bounding box
- limits X and Y to the visible area in the Image View panel
- restores full dataset dimensions
- applies a binning factor to reduce detail for faster processing
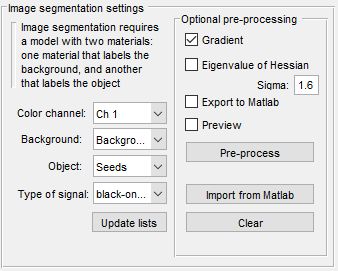
Image segmentation settings
The Watershed workflow uses labeled areas (Background and Object) for segmentation, contrasting with the Graphcut workflow. It’s less interactive, slower per execution, and best for objects with distinct boundaries. Graphcut, with its preprocessing focus, is faster for repeated interactions and handles both boundaries and intensity contrast, making it preferable for most cases.
- selects the channel for segmentation
- selects the model material labeling background areas
- selects the model material labeling objects
- chooses black-on-white (dark boundaries) or white-on-black (bright boundaries)
- refreshes material lists
- applies a gradient filter to enhance object borders
- preprocesses data for improved Watershed results; adjust with Sigma fields
- sends preprocessed data to the MATLAB workspace
- displays preprocessing results in the Image View panel
- starts preprocessing (turns green when data is ready)
- loads a dataset from the MATLAB workspace for segmentation
- removes preprocessed data from memory
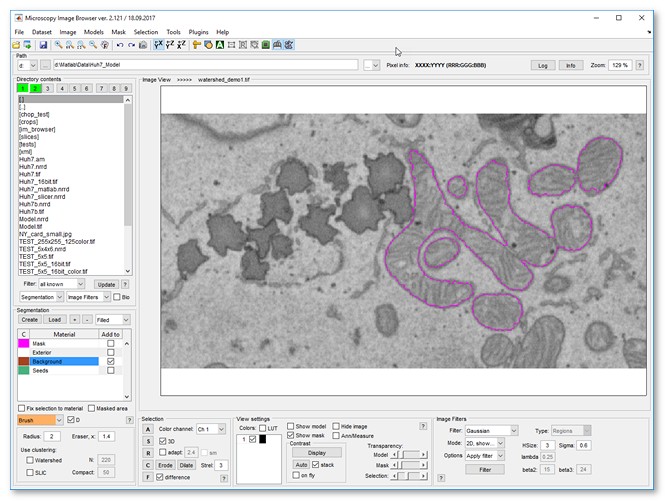
Image segmentation example
Follow these steps to segment mitochondria using Watershed:
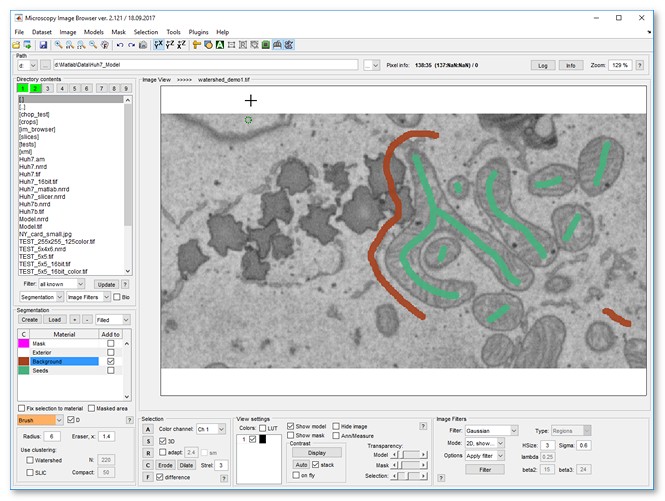
- Load a sample dataset:
Menu → File → Import image from → URL, enter
http://mib.helsinki.fi/tutorials/WatershedDemo/watershed_demo1.tif - Add a material named Background in the Segmentation panel with (right-click to rename)
- Use the Brush tool to label cytoplasm, then press to add it to Background
- Add another material named Seeds with
- Label mitochondria interiors, then press to add to Seeds
- Start Watershed via
Menu → Tools → Semi-automatic segmentation → Watershed - Ensure Background and Seeds are set in Image segmentation settings
- Click to segment mitochondria
- Add more seeds to refine, then re-segment with
- Results appear in the Mask layer
- Optionally smooth the mask: Menu → Mask → Smooth Mask
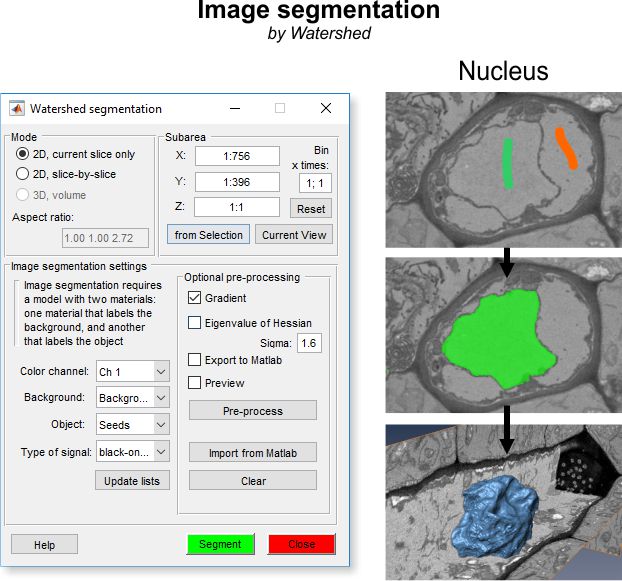
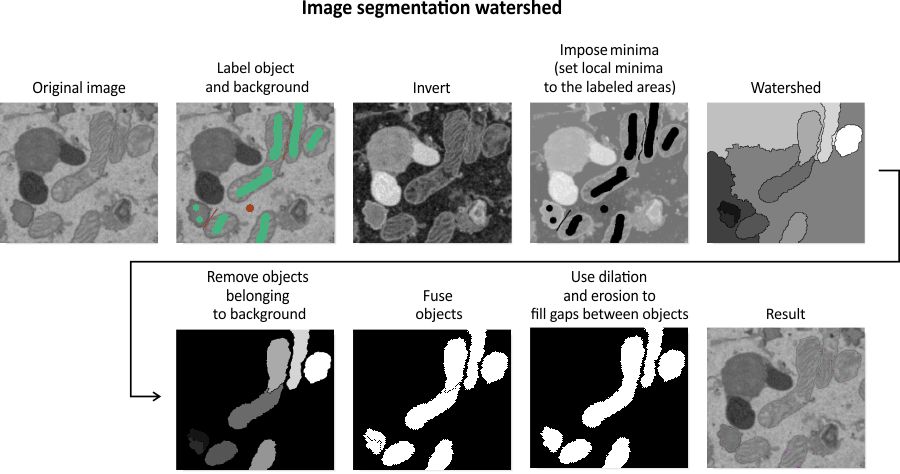
Algorithm for image segmentation with watershed
The diagram illustrates the step-by-step process of the Watershed segmentation algorithm, transforming labeled input into segmented objects.
References
- Watershed transform question from tech support by Steve Eddins
- Cell segmentation by Steve Eddins
Back to MIB | User interface | Menu | Tools