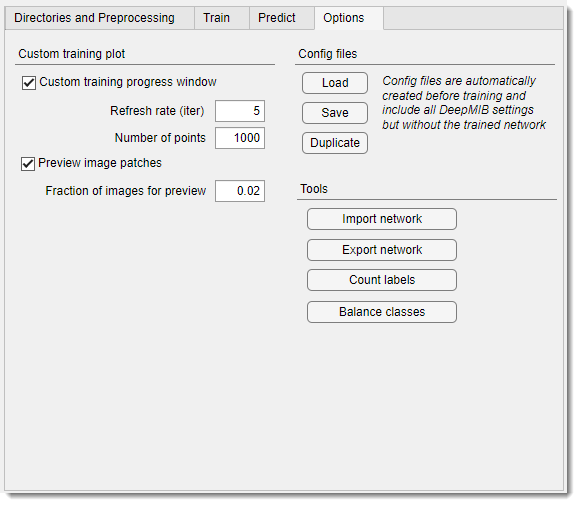
Deep MIB - Options Tab
Back to MIB | User interface | DeepMIB
Additional options and settings for deep learning segmentation in Microscopy Image Browser.
Overview
The Options tab provides supplementary settings for customizing Deep MIB’s behavior during training, prediction, and data management.
Custom training plot section
Configures the custom training progress plot displaying the loss function during training.
enables the custom plot when checked; unchecked uses MATLAB’s standard plot (MATLAB version only)
sets the iteration interval for plot updates. Higher values reduce updates, boosting training performance
defines the number of plot points. Lower values enhance drawing speed, higher values show more detail
displays input image and model patches in the custom plot, reducing performance.
Adjust visibility with
sets the fraction of patches shown (1 = all, 0.01 = 1%)
Config files section
Manages Deep MIB configuration files (.mibCfg), which store all settings, including network name and directories, but not the trained network itself. these are auto-generated during training alongside .mibDeep files or saved manually.
- loads DeepMIB config file (
.mibCfg) to restore all settings - manually saves the current configuration
- copies the network (
.mibDeep) and config (.mibCfg) to new files, updating the network name in the config. This operation is useful for backups or when another network needs be trained from already trained one.
Tools section
Import network
imports externally designed or trained networks (MATLAB format only).
Use MATLAB Deep Network Designer to create networks, then import them into Deep MIB for training or prediction,
generating .mibCfg and .mibDeep files.
Export network
exports trained networks to ONNX or TensorFlow formats.
Additional details of the export process
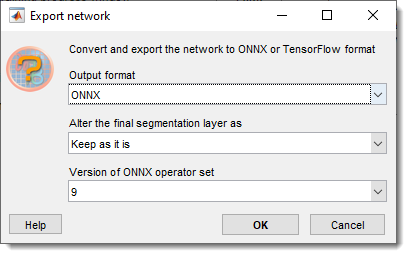
- : chooses ONNX operator set (6, 7, 8, 9)
- : modifies the last segmentation layer (e.g., replacing non-standard layers like CustomDice for ONNX compatibility)
list of available options
- Keep as it is: retains the original layer
- Remove the layer: removes the segmentation layer, making softmax the final layer
- pixelClassificationLayer: swaps to a standard pixel classification layer
- dicePixelClassificationLayer: swaps to a Dice loss-based layer
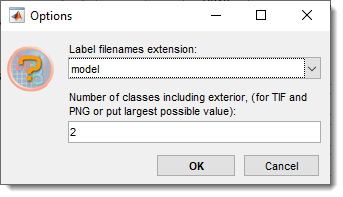
Count labels
counts labels in model files.
Details of Count Labels
- Select the directory with labels
- Specify the file extension (
.model,.mibCat,.png,.tif,.tiff) and number of classes
- Save results to
.mat,.xls, or.csv
Balance classes
(beta) balances rare classes in 2D Semantic workflows for standard image formats (TIF, PNG, JPG), generating patches from large rasters.
Details of Balance classes
Place images and labels in Images and Labels subfolders under
Directories and Preprocessing → Directory with images and labels for training.
Balanced results are saved to ImagesBalanced and LabelsBalanced subfolders.
Driven by MATLAB’s balancePixelLabels function.
Back to MIB | User interface | DeepMIB