Surface Area 3D
Back to MIB | User Interface | Plugins | Organelle Analysis
The Surface Area 3D plugin in Microscopy Image Browser (MIB) calculates planar surface areas of 3D objects in microscopy datasets, such as organelle contacts (e.g., mitochondria and ER). It detects objects using 26-point connectivity, computes surface areas from centerlines, and supports visualization and export to various formats.
Overview
The plugin analyzes 3D objects by generating triangulated surfaces from cross-sectional centerlines.
Key features include:
- Detection of 3D objects with 26-edge connectivity.
- Calculation of surface areas per object.
- Export to MATLAB, Imaris, Amira, CSV, Excel, or MATLAB binary formats.
- Visualization of surfaces in MATLAB or MIB’s
Selectionlayer.
Parameters
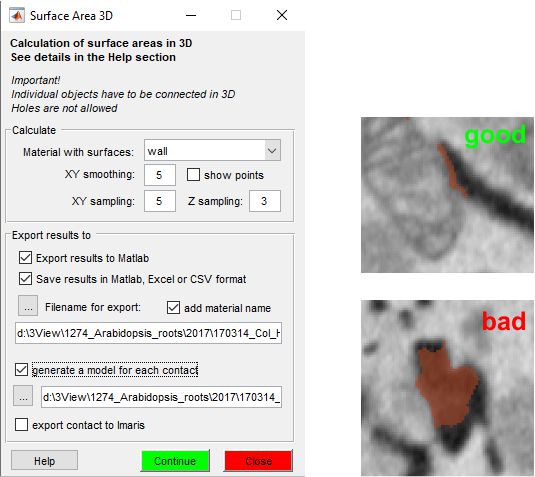
Configure the analysis with these settings:
- select the model material to analyze. Objects are detected in 3D with 26-edge connectivity, and surfaces are generated per object.
- smooth centerline points’ coordinates (XY plane) to refine profiles.
- increase triangle size by skipping points (e.g., 3 uses every third point for triangulation).
- similar to XY sampling, applied to Z-dimension.
- display detected points in the Selection layer. Clear afterward with Shift+C to avoid model conflicts.
- generate a structure in the main MATLAB workspace.
Configiration of the export structure
.pixSize: dataset pixel size (SurfaceArea(1).pixSize)..MaterialName: analyzed material name (SurfaceArea(1).MaterialName)..xySmoothValue: XY smoothing factor..xySamplingStep: XY sampling value..zSamplingStep: Z sampling value..SumAreaTotal: total area per surface (SurfaceArea(N).SumAreaTotal)..PointsVector: point indices per profile ([x,y,z])..PointCloud: all points as[index](x,y,z)..Centroid: object centroids..Area: area between consecutive slices (cell array).
- save to MATLAB, Excel, or CSV.
- append material name to
output filenames. - save surfaces in AmiraMesh format to a folder.
- export surfaces to Imaris (requires Imaris 8 and ImarisXT).
Warning
Clear the Selection layer after checking points (Shift+C) to prevent unintended
changes to the model.
Usage
Access the plugin via:
Menu → Plugins → Organelle Analysis → Surface Area 3D.
The following steps outline analysis, using an example of mitochondria-ER contacts:
Load Dataset and Model:
- Open a 3D dataset (e.g., Huh7.tif).
- Load a model with materials (e.g., Labels_Huh.model) via
Menu → File → Load model.
Open from Menu
It is possible to load both the dataset and the model using
Menu->File->Example datasets->SBEM->Huh7 and model (29 Mb) (see more)
Help Resources
- Tutorial Video: Surface Area 3D Demo (Note: Video refers to plugin as ContactArea3D).
- External Tutorial: Plasmodesmata Analysis.
Credits and Acknowledgements
Author: Ilya Belevich, University of Helsinki
- Version: 1.00, 13.02.2020
- Email: ilya.belevich@helsinki.fi
Big thanks to David Legland (Institut National de la Recherche Agronomique, France)
for discussion about triangulation of points:
function mibTriangulateCurvePair.m is based on triangulateCurvePair.m from MatGeom tools
by David Legland
How to Cite
If you use this plugin, please check for citation:
Data analysis pipeline in Paterlini, Belevich et al., 2020
Back to MIB | User Interface | Plugins | Organelle Analysis